Wenrui Fan
Classifying the Stoichiometry of Virus-like Particles with Interpretable Machine Learning
Feb 17, 2025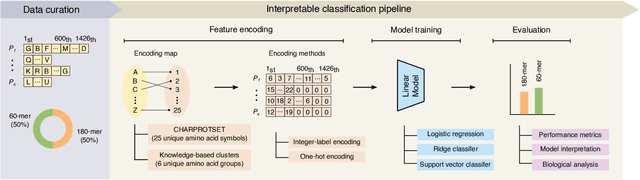
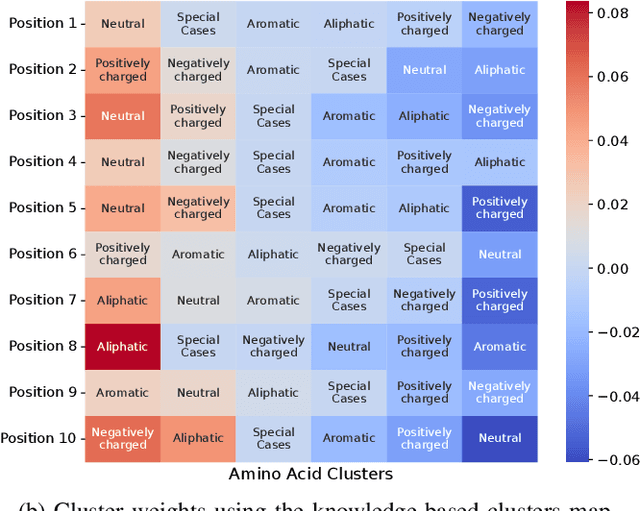
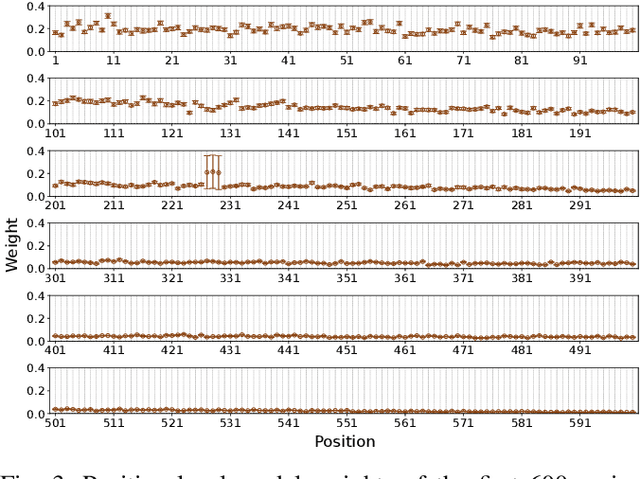
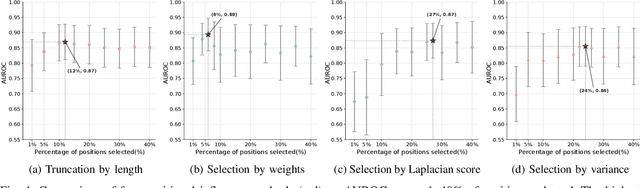
Abstract:Virus-like particles (VLPs) are valuable for vaccine development due to their immune-triggering properties. Understanding their stoichiometry, the number of protein subunits to form a VLP, is critical for vaccine optimisation. However, current experimental methods to determine stoichiometry are time-consuming and require highly purified proteins. To efficiently classify stoichiometry classes in proteins, we curate a new dataset and propose an interpretable, data-driven pipeline leveraging linear machine learning models. We also explore the impact of feature encoding on model performance and interpretability, as well as methods to identify key protein sequence features influencing classification. The evaluation of our pipeline demonstrates that it can classify stoichiometry while revealing protein features that possibly influence VLP assembly. The data and code used in this work are publicly available at https://github.com/Shef-AIRE/StoicIML.
Multimodal Variational Autoencoder for Low-cost Cardiac Hemodynamics Instability Detection
Mar 20, 2024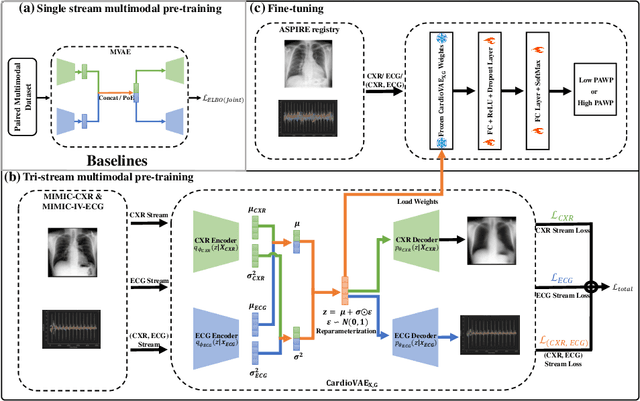
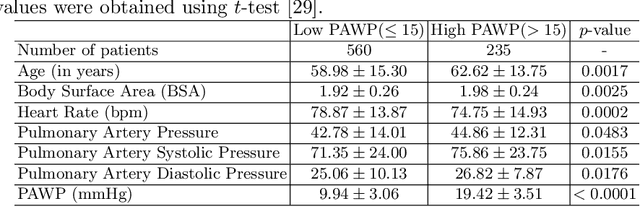
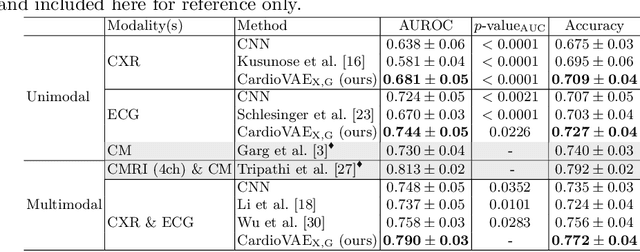
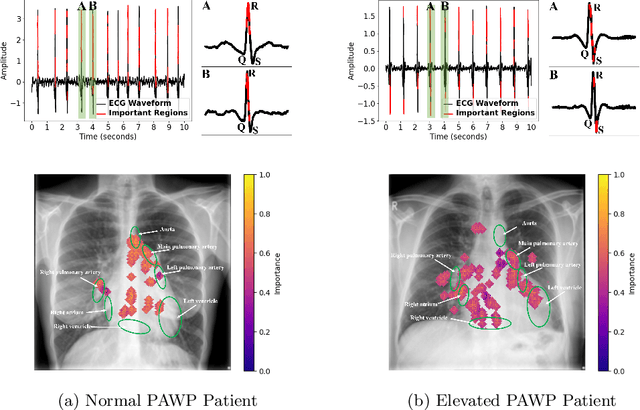
Abstract:Recent advancements in non-invasive detection of cardiac hemodynamic instability (CHDI) primarily focus on applying machine learning techniques to a single data modality, e.g. cardiac magnetic resonance imaging (MRI). Despite their potential, these approaches often fall short especially when the size of labeled patient data is limited, a common challenge in the medical domain. Furthermore, only a few studies have explored multimodal methods to study CHDI, which mostly rely on costly modalities such as cardiac MRI and echocardiogram. In response to these limitations, we propose a novel multimodal variational autoencoder ($\text{CardioVAE}_\text{X,G}$) to integrate low-cost chest X-ray (CXR) and electrocardiogram (ECG) modalities with pre-training on a large unlabeled dataset. Specifically, $\text{CardioVAE}_\text{X,G}$ introduces a novel tri-stream pre-training strategy to learn both shared and modality-specific features, thus enabling fine-tuning with both unimodal and multimodal datasets. We pre-train $\text{CardioVAE}_\text{X,G}$ on a large, unlabeled dataset of $50,982$ subjects from a subset of MIMIC database and then fine-tune the pre-trained model on a labeled dataset of $795$ subjects from the ASPIRE registry. Comprehensive evaluations against existing methods show that $\text{CardioVAE}_\text{X,G}$ offers promising performance (AUROC $=0.79$ and Accuracy $=0.77$), representing a significant step forward in non-invasive prediction of CHDI. Our model also excels in producing fine interpretations of predictions directly associated with clinical features, thereby supporting clinical decision-making.
MeDSLIP: Medical Dual-Stream Language-Image Pre-training for Fine-grained Alignment
Mar 15, 2024Abstract:Vision-language pre-training (VLP) models have shown significant advancements in the medical domain. Yet, most VLP models align raw reports to images at a very coarse level, without modeling fine-grained relationships between anatomical and pathological concepts outlined in reports and the corresponding semantic counterparts in images. To address this problem, we propose a Medical Dual-Stream Language-Image Pre-training (MeDSLIP) framework. Specifically, MeDSLIP establishes vision-language fine-grained alignments via disentangling visual and textual representations into anatomy-relevant and pathology-relevant streams. Moreover, a novel vision-language Prototypical Contr-astive Learning (ProtoCL) method is adopted in MeDSLIP to enhance the alignment within the anatomical and pathological streams. MeDSLIP further employs cross-stream Intra-image Contrastive Learning (ICL) to ensure the consistent coexistence of paired anatomical and pathological concepts within the same image. Such a cross-stream regularization encourages the model to exploit the synchrony between two streams for a more comprehensive representation learning. MeDSLIP is evaluated under zero-shot and supervised fine-tuning settings on three public datasets: NIH CXR14, RSNA Pneumonia, and SIIM-ACR Pneumothorax. Under these settings, MeDSLIP outperforms six leading CNN-based models on classification, grounding, and segmentation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge