Andrew J. Swift
CMRINet: Joint Groupwise Registration and Segmentation for Cardiac Function Quantification from Cine-MRI
May 22, 2025Abstract:Accurate and efficient quantification of cardiac function is essential for the estimation of prognosis of cardiovascular diseases (CVDs). One of the most commonly used metrics for evaluating cardiac pumping performance is left ventricular ejection fraction (LVEF). However, LVEF can be affected by factors such as inter-observer variability and varying pre-load and after-load conditions, which can reduce its reproducibility. Additionally, cardiac dysfunction may not always manifest as alterations in LVEF, such as in heart failure and cardiotoxicity diseases. An alternative measure that can provide a relatively load-independent quantitative assessment of myocardial contractility is myocardial strain and strain rate. By using LVEF in combination with myocardial strain, it is possible to obtain a thorough description of cardiac function. Automated estimation of LVEF and other volumetric measures from cine-MRI sequences can be achieved through segmentation models, while strain calculation requires the estimation of tissue displacement between sequential frames, which can be accomplished using registration models. These tasks are often performed separately, potentially limiting the assessment of cardiac function. To address this issue, in this study we propose an end-to-end deep learning (DL) model that jointly estimates groupwise (GW) registration and segmentation for cardiac cine-MRI images. The proposed anatomically-guided Deep GW network was trained and validated on a large dataset of 4-chamber view cine-MRI image series of 374 subjects. A quantitative comparison with conventional GW registration using elastix and two DL-based methods showed that the proposed model improved performance and substantially reduced computation time.
TabMixer: Noninvasive Estimation of the Mean Pulmonary Artery Pressure via Imaging and Tabular Data Mixing
Sep 11, 2024Abstract:Right Heart Catheterization is a gold standard procedure for diagnosing Pulmonary Hypertension by measuring mean Pulmonary Artery Pressure (mPAP). It is invasive, costly, time-consuming and carries risks. In this paper, for the first time, we explore the estimation of mPAP from videos of noninvasive Cardiac Magnetic Resonance Imaging. To enhance the predictive capabilities of Deep Learning models used for this task, we introduce an additional modality in the form of demographic features and clinical measurements. Inspired by all-Multilayer Perceptron architectures, we present TabMixer, a novel module enabling the integration of imaging and tabular data through spatial, temporal and channel mixing. Specifically, we present the first approach that utilizes Multilayer Perceptrons to interchange tabular information with imaging features in vision models. We test TabMixer for mPAP estimation and show that it enhances the performance of Convolutional Neural Networks, 3D-MLP and Vision Transformers while being competitive with previous modules for imaging and tabular data. Our approach has the potential to improve clinical processes involving both modalities, particularly in noninvasive mPAP estimation, thus, significantly enhancing the quality of life for individuals affected by Pulmonary Hypertension. We provide a source code for using TabMixer at https://github.com/SanoScience/TabMixer.
Multimodal Variational Autoencoder for Low-cost Cardiac Hemodynamics Instability Detection
Mar 20, 2024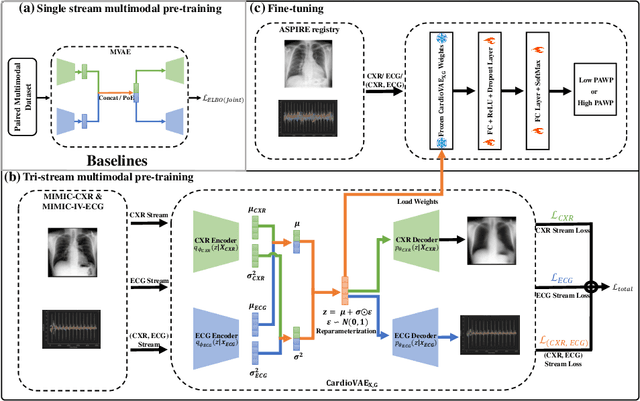
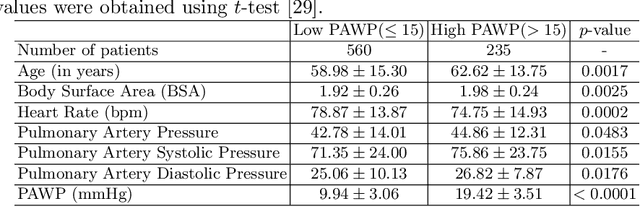
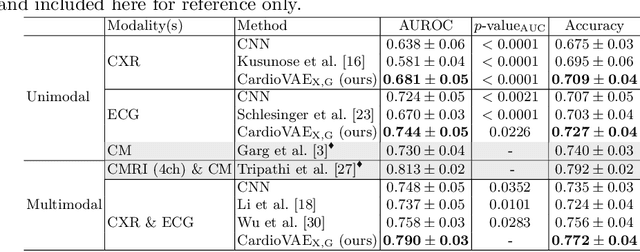
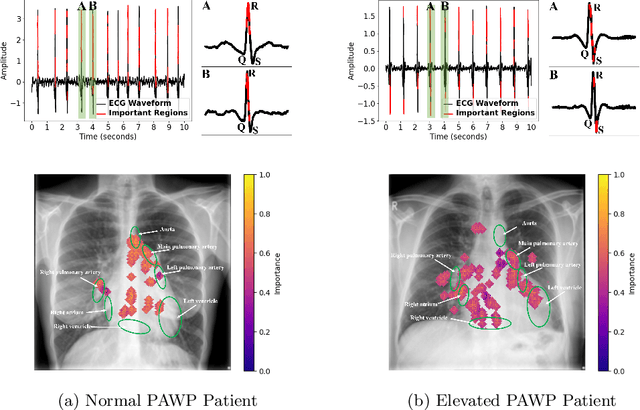
Abstract:Recent advancements in non-invasive detection of cardiac hemodynamic instability (CHDI) primarily focus on applying machine learning techniques to a single data modality, e.g. cardiac magnetic resonance imaging (MRI). Despite their potential, these approaches often fall short especially when the size of labeled patient data is limited, a common challenge in the medical domain. Furthermore, only a few studies have explored multimodal methods to study CHDI, which mostly rely on costly modalities such as cardiac MRI and echocardiogram. In response to these limitations, we propose a novel multimodal variational autoencoder ($\text{CardioVAE}_\text{X,G}$) to integrate low-cost chest X-ray (CXR) and electrocardiogram (ECG) modalities with pre-training on a large unlabeled dataset. Specifically, $\text{CardioVAE}_\text{X,G}$ introduces a novel tri-stream pre-training strategy to learn both shared and modality-specific features, thus enabling fine-tuning with both unimodal and multimodal datasets. We pre-train $\text{CardioVAE}_\text{X,G}$ on a large, unlabeled dataset of $50,982$ subjects from a subset of MIMIC database and then fine-tune the pre-trained model on a labeled dataset of $795$ subjects from the ASPIRE registry. Comprehensive evaluations against existing methods show that $\text{CardioVAE}_\text{X,G}$ offers promising performance (AUROC $=0.79$ and Accuracy $=0.77$), representing a significant step forward in non-invasive prediction of CHDI. Our model also excels in producing fine interpretations of predictions directly associated with clinical features, thereby supporting clinical decision-making.
Deep learning automated quantification of lung disease in pulmonary hypertension on CT pulmonary angiography: A preliminary clinical study with external validation
Mar 20, 2023



Abstract:Purpose: Lung disease assessment in precapillary pulmonary hypertension (PH) is essential for appropriate patient management. This study aims to develop an artificial intelligence (AI) deep learning model for lung texture classification in CT Pulmonary Angiography (CTPA), and evaluate its correlation with clinical assessment methods. Materials and Methods: In this retrospective study with external validation, 122 patients with pre-capillary PH were used to train (n=83), validate (n=17) and test (n=10 internal test, n=12 external test) a patch based DenseNet-121 classification model. "Normal", "Ground glass", "Ground glass with reticulation", "Honeycombing", and "Emphysema" were classified as per the Fleishner Society glossary of terms. Ground truth classes were segmented by two radiologists with patches extracted from the labelled regions. Proportion of lung volume for each texture was calculated by classifying patches throughout the entire lung volume to generate a coarse texture classification mapping throughout the lung parenchyma. AI output was assessed against diffusing capacity of carbon monoxide (DLCO) and specialist radiologist reported disease severity. Results: Micro-average AUCs for the validation, internal test, and external test were 0.92, 0.95, and 0.94, respectively. The model had consistent performance across parenchymal textures, demonstrated strong correlation with diffusing capacity of carbon monoxide (DLCO), and showed good correspondence with disease severity reported by specialist radiologists. Conclusion: The classification model demonstrates excellent performance on external validation. The clinical utility of its output has been demonstrated. This objective, repeatable measure of disease severity can aid in patient management in adjunct to radiological reporting.
Tensor-based Multimodal Learning for Prediction of Pulmonary Arterial Wedge Pressure from Cardiac MRI
Mar 14, 2023Abstract:Heart failure is a serious and life-threatening condition that can lead to elevated pressure in the left ventricle. Pulmonary Arterial Wedge Pressure (PAWP) is an important surrogate marker indicating high pressure in the left ventricle. PAWP is determined by Right Heart Catheterization (RHC) but it is an invasive procedure. A non-invasive method is useful in quickly identifying high-risk patients from a large population. In this work, we develop a tensor learning-based pipeline for identifying PAWP from multimodal cardiac Magnetic Resonance Imaging (MRI). This pipeline extracts spatial and temporal features from high-dimensional scans. For quality control, we incorporate an epistemic uncertainty-based binning strategy to identify poor-quality training samples. To improve the performance, we learn complementary information by integrating features from multimodal data: cardiac MRI with short-axis and four-chamber views, and Electronic Health Records. The experimental analysis on a large cohort of $1346$ subjects who underwent the RHC procedure for PAWP estimation indicates that the proposed pipeline has a diagnostic value and can produce promising performance with significant improvement over the baseline in clinical practice (i.e., $\Delta$AUC $=0.10$, $\Delta$Accuracy $=0.06$, and $\Delta$MCC $=0.39$). The decision curve analysis further confirms the clinical utility of our method.
Uncertainty Estimation for Heatmap-based Landmark Localization
Mar 04, 2022



Abstract:Automatic anatomical landmark localization has made great strides by leveraging deep learning methods in recent years. The ability to quantify the uncertainty of these predictions is a vital ingredient needed to see these methods adopted in clinical use, where it is imperative that erroneous predictions are caught and corrected. We propose Quantile Binning, a data-driven method to categorise predictions by uncertainty with estimated error bounds. This framework can be applied to any continuous uncertainty measure, allowing straightforward identification of the best subset of predictions with accompanying estimated error bounds. We facilitate easy comparison between uncertainty measures by constructing two evaluation metrics derived from Quantile Binning. We demonstrate this framework by comparing and contrasting three uncertainty measures (a baseline, the current gold standard, and a proposed method combining aspects of the two), across two datasets (one easy, one hard) and two heatmap-based landmark localization model paradigms (U-Net and patch-based). We conclude by illustrating how filtering out gross mispredictions caught in our Quantile Bins significantly improves the proportion of predictions under an acceptable error threshold, and offer recommendations on which uncertainty measure to use and how to use it.
DenResCov-19: A deep transfer learning network for robust automatic classification of COVID-19, pneumonia, and tuberculosis from X-rays
Apr 08, 2021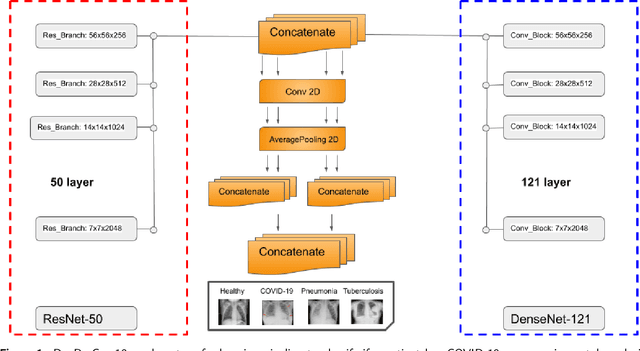
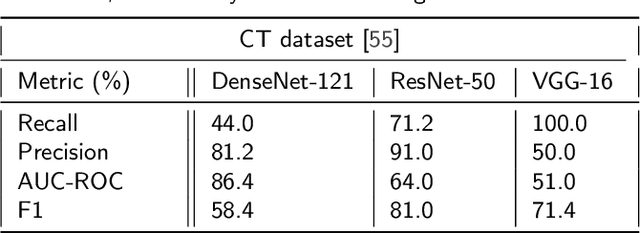
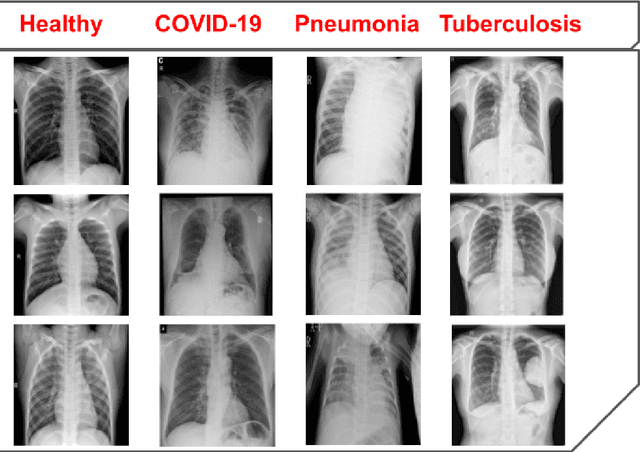
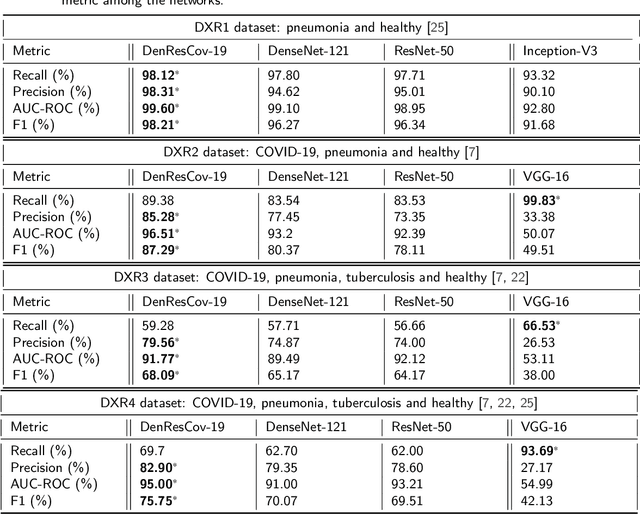
Abstract:The global pandemic of COVID-19 is continuing to have a significant effect on the well-being of global population, increasing the demand for rapid testing, diagnosis, and treatment. Along with COVID-19, other etiologies of pneumonia and tuberculosis constitute additional challenges to the medical system. In this regard, the objective of this work is to develop a new deep transfer learning pipeline to diagnose patients with COVID-19, pneumonia, and tuberculosis, based on chest x-ray images. We observed in some instances DenseNet and Resnet have orthogonal performances. In our proposed model, we have created an extra layer with convolutional neural network blocks to combine these two models to establish superior performance over either model. The same strategy can be useful in other applications where two competing networks with complementary performance are observed. We have tested the performance of our proposed network on two-class (pneumonia vs healthy), three-class (including COVID-19), and four-class (including tuberculosis) classification problems. The proposed network has been able to successfully classify these lung diseases in all four datasets and has provided significant improvement over the benchmark networks of DenseNet, ResNet, and Inception-V3. These novel findings can deliver a state-of-the-art pre-screening fast-track decision network to detect COVID-19 and other lung pathologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge