Abhirup Banerjee
From 2D to 3D, Deep Learning-based Shape Reconstruction in Magnetic Resonance Imaging: A Review
Oct 01, 2025Abstract:Deep learning-based 3-dimensional (3D) shape reconstruction from 2-dimensional (2D) magnetic resonance imaging (MRI) has become increasingly important in medical disease diagnosis, treatment planning, and computational modeling. This review surveys the methodological landscape of 3D MRI reconstruction, focusing on 4 primary approaches: point cloud, mesh-based, shape-aware, and volumetric models. For each category, we analyze the current state-of-the-art techniques, their methodological foundation, limitations, and applications across anatomical structures. We provide an extensive overview ranging from cardiac to neurological to lung imaging. We also focus on the clinical applicability of models to diseased anatomy, and the influence of their training and testing data. We examine publicly available datasets, computational demands, and evaluation metrics. Finally, we highlight the emerging research directions including multimodal integration and cross-modality frameworks. This review aims to provide researchers with a structured overview of current 3D reconstruction methodologies to identify opportunities for advancing deep learning towards more robust, generalizable, and clinically impactful solutions.
NeCA: 3D Coronary Artery Tree Reconstruction from Two 2D Projections by Neural Implicit Representation
Sep 06, 2024Abstract:Cardiovascular diseases (CVDs) are the most common health threats worldwide. 2D x-ray invasive coronary angiography (ICA) remains as the most widely adopted imaging modality for CVDs diagnosis. However, in current clinical practice, it is often difficult for the cardiologists to interpret the 3D geometry of coronary vessels based on 2D planes. Moreover, due to the radiation limit, in general only two angiographic projections are acquired, providing limited information of the vessel geometry and necessitating 3D coronary tree reconstruction based only on two ICA projections. In this paper, we propose a self-supervised deep learning method called NeCA, which is based on implicit neural representation using the multiresolution hash encoder and differentiable cone-beam forward projector layer in order to achieve 3D coronary artery tree reconstruction from two projections. We validate our method using six different metrics on coronary computed tomography angiography data in terms of right coronary artery and left anterior descending respectively. The evaluation results demonstrate that our NeCA method, without 3D ground truth for supervision and large datasets for training, achieves promising performance in both vessel topology preservation and branch-connectivity maintaining compared to the supervised deep learning model.
Personalized Topology-Informed 12-Lead ECG Electrode Localization from Incomplete Cardiac MRIs for Efficient Cardiac Digital Twins
Aug 25, 2024Abstract:Cardiac digital twins (CDTs) offer personalized \textit{in-silico} cardiac representations for the inference of multi-scale properties tied to cardiac mechanisms. The creation of CDTs requires precise information about the electrode position on the torso, especially for the personalized electrocardiogram (ECG) calibration. However, current studies commonly rely on additional acquisition of torso imaging and manual/semi-automatic methods for ECG electrode localization. In this study, we propose a novel and efficient topology-informed model to fully automatically extract personalized ECG electrode locations from 2D clinically standard cardiac MRIs. Specifically, we obtain the sparse torso contours from the cardiac MRIs and then localize the electrodes from the contours. Cardiac MRIs aim at imaging of the heart instead of the torso, leading to incomplete torso geometry within the imaging. To tackle the missing topology, we incorporate the electrodes as a subset of the keypoints, which can be explicitly aligned with the 3D torso topology. The experimental results demonstrate that the proposed model outperforms the time-consuming conventional method in terms of accuracy (Euclidean distance: $1.24 \pm 0.293$ cm vs. $1.48 \pm 0.362$ cm) and efficiency ($2$~s vs. $30$-$35$~min). We further demonstrate the effectiveness of using the detected electrodes for \textit{in-silico} ECG simulation, highlighting their potential for creating accurate and efficient CDT models. The code will be released publicly after the manuscript is accepted for publication.
Deep Learning-based 3D Coronary Tree Reconstruction from Two 2D Non-simultaneous X-ray Angiography Projections
Jul 19, 2024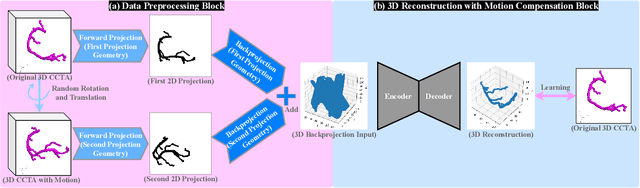

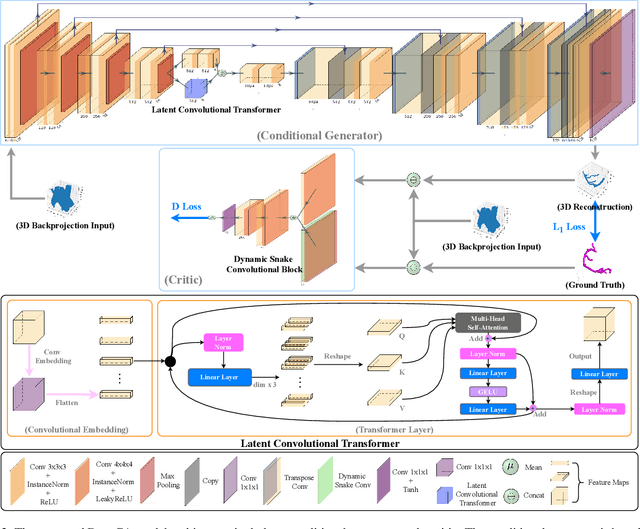

Abstract:Cardiovascular diseases (CVDs) are the most common cause of death worldwide. Invasive x-ray coronary angiography (ICA) is one of the most important imaging modalities for the diagnosis of CVDs. ICA typically acquires only two 2D projections, which makes the 3D geometry of coronary vessels difficult to interpret, thus requiring 3D coronary tree reconstruction from two projections. State-of-the-art approaches require significant manual interactions and cannot correct the non-rigid cardiac and respiratory motions between non-simultaneous projections. In this study, we propose a novel deep learning pipeline. We leverage the Wasserstein conditional generative adversarial network with gradient penalty, latent convolutional transformer layers, and a dynamic snake convolutional critic to implicitly compensate for the non-rigid motion and provide 3D coronary tree reconstruction. Through simulating projections from coronary computed tomography angiography (CCTA), we achieve the generalisation of 3D coronary tree reconstruction on real non-simultaneous ICA projections. We incorporate an application-specific evaluation metric to validate our proposed model on both a CCTA dataset and a real ICA dataset, together with Chamfer L1 distance. The results demonstrate the good performance of our model in vessel topology preservation, recovery of missing features, and generalisation ability to real ICA data. To the best of our knowledge, this is the first study that leverages deep learning to achieve 3D coronary tree reconstruction from two real non-simultaneous x-ray angiography projections.
Feasibility and benefits of joint learning from MRI databases with different brain diseases and modalities for segmentation
May 28, 2024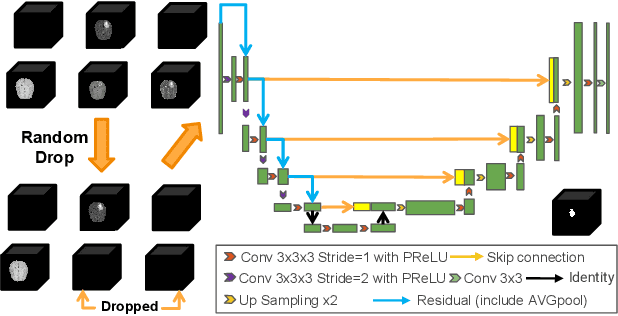
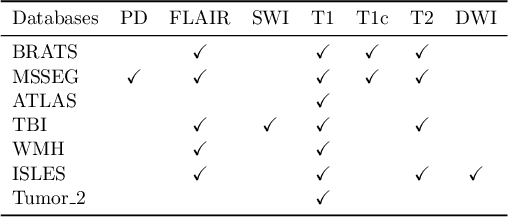
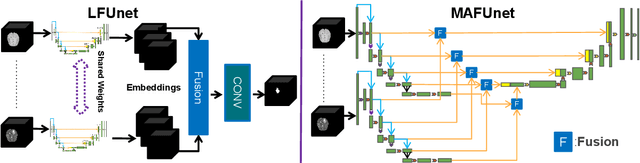

Abstract:Models for segmentation of brain lesions in multi-modal MRI are commonly trained for a specific pathology using a single database with a predefined set of MRI modalities, determined by a protocol for the specific disease. This work explores the following open questions: Is it feasible to train a model using multiple databases that contain varying sets of MRI modalities and annotations for different brain pathologies? Will this joint learning benefit performance on the sets of modalities and pathologies available during training? Will it enable analysis of new databases with different sets of modalities and pathologies? We develop and compare different methods and show that promising results can be achieved with appropriate, simple and practical alterations to the model and training framework. We experiment with 7 databases containing 5 types of brain pathologies and different sets of MRI modalities. Results demonstrate, for the first time, that joint training on multi-modal MRI databases with different brain pathologies and sets of modalities is feasible and offers practical benefits. It enables a single model to segment pathologies encountered during training in diverse sets of modalities, while facilitating segmentation of new types of pathologies such as via follow-up fine-tuning. The insights this study provides into the potential and limitations of this paradigm should prove useful for guiding future advances in the direction. Code and pretrained models: https://github.com/WenTXuL/MultiUnet
* Accepted to MIDL 2024
Large Language Model-informed ECG Dual Attention Network for Heart Failure Risk Prediction
Mar 22, 2024
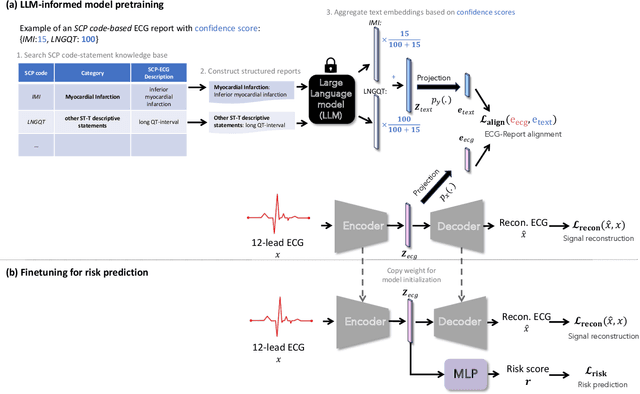
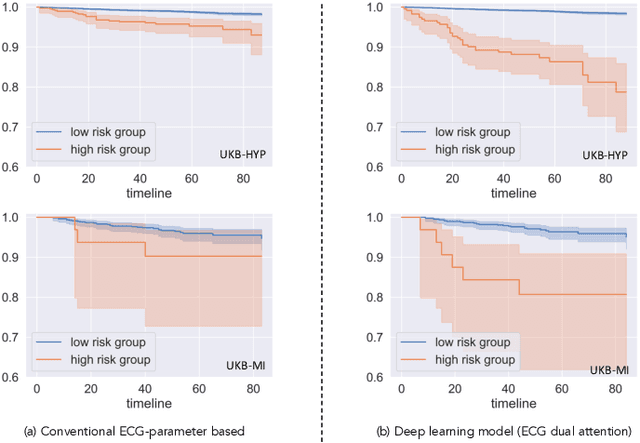
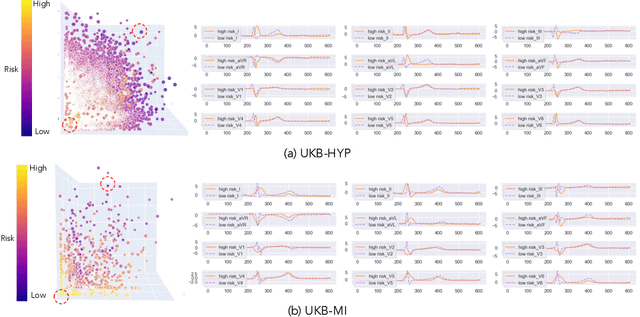
Abstract:Heart failure (HF) poses a significant public health challenge, with a rising global mortality rate. Early detection and prevention of HF could significantly reduce its impact. We introduce a novel methodology for predicting HF risk using 12-lead electrocardiograms (ECGs). We present a novel, lightweight dual-attention ECG network designed to capture complex ECG features essential for early HF risk prediction, despite the notable imbalance between low and high-risk groups. This network incorporates a cross-lead attention module and twelve lead-specific temporal attention modules, focusing on cross-lead interactions and each lead's local dynamics. To further alleviate model overfitting, we leverage a large language model (LLM) with a public ECG-Report dataset for pretraining on an ECG-report alignment task. The network is then fine-tuned for HF risk prediction using two specific cohorts from the UK Biobank study, focusing on patients with hypertension (UKB-HYP) and those who have had a myocardial infarction (UKB-MI).The results reveal that LLM-informed pre-training substantially enhances HF risk prediction in these cohorts. The dual-attention design not only improves interpretability but also predictive accuracy, outperforming existing competitive methods with C-index scores of 0.6349 for UKB-HYP and 0.5805 for UKB-MI. This demonstrates our method's potential in advancing HF risk assessment with clinical complex ECG data.
Anatomical basis of sex differences in human post-myocardial infarction ECG phenotypes identified by novel automated torso-cardiac 3D reconstruction
Dec 21, 2023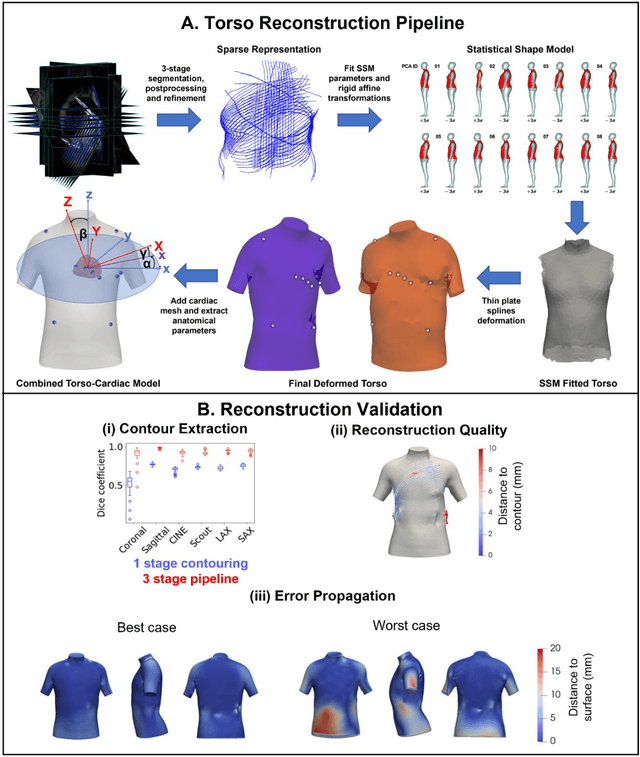
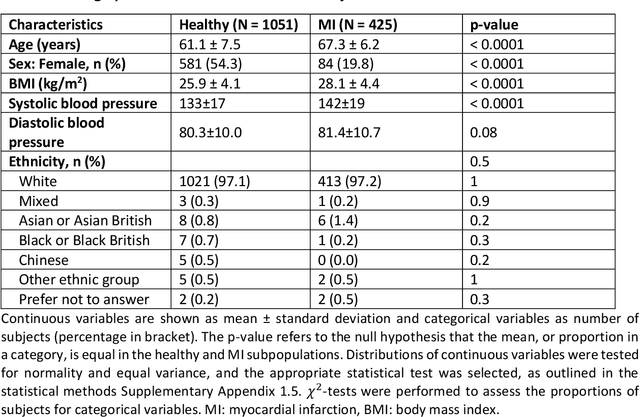
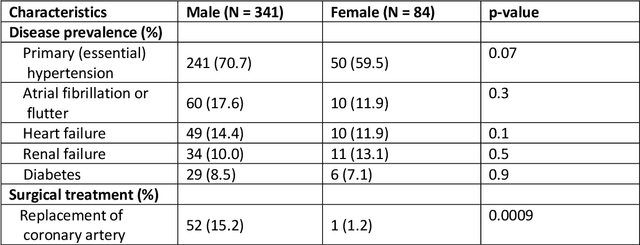
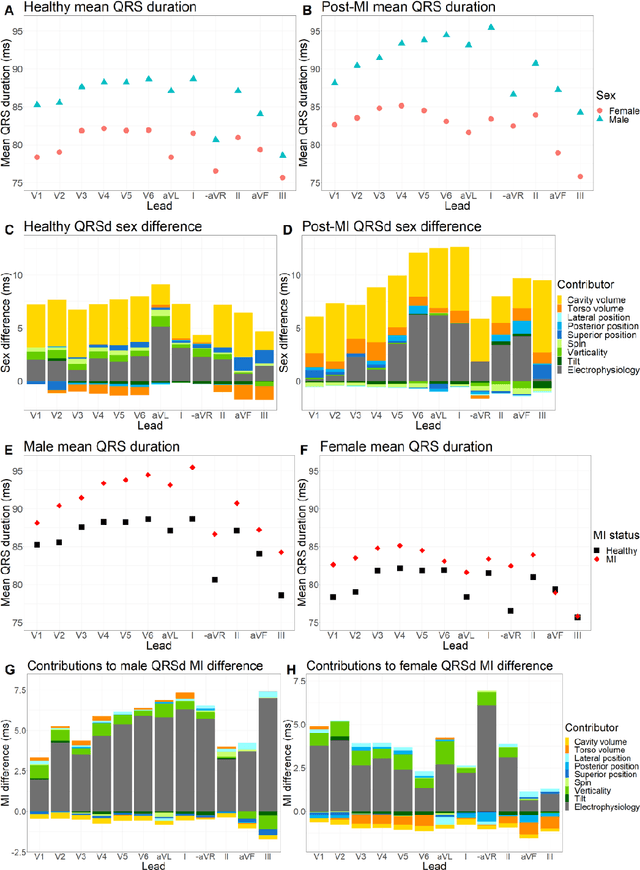
Abstract:The electrocardiogram (ECG) is routinely used in cardiology, though its interpretation is confounded by anatomical variability. A novel, automated computational pipeline enables quantification of torso-ventricular anatomy metrics from magnetic resonance imaging, and comparison to ECG characteristics. Sex and myocardial infarction differences are investigated based on 1051 healthy and 425 post-MI subjects from UK Biobank. Smaller ventricles in females explain ~50% of shorter QRS durations than in males, and contribute to lower STJ amplitudes in females (also due to more superior and posterior position). In females, torso-ventricular anatomy, particularly from larger BMI, is a stronger modulator of T wave amplitude reductions and left-deviated R axis angles in post-MI than in males. Thus, female MI phenotype is less reflective of pathology, and baseline STJ amplitudes and QRS durations are further from clinical thresholds. Therefore, quantification of anatomical sex-differences and impact on ECG in health and disease is critical to avoid clinical sex-bias.
Hunting imaging biomarkers in pulmonary fibrosis: Benchmarks of the AIIB23 challenge
Dec 21, 2023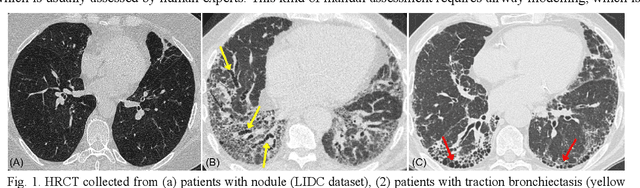
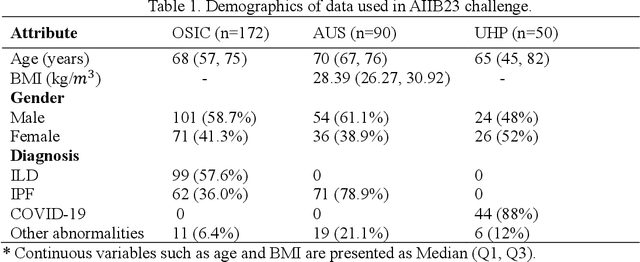
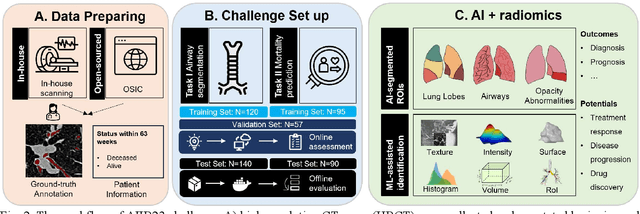
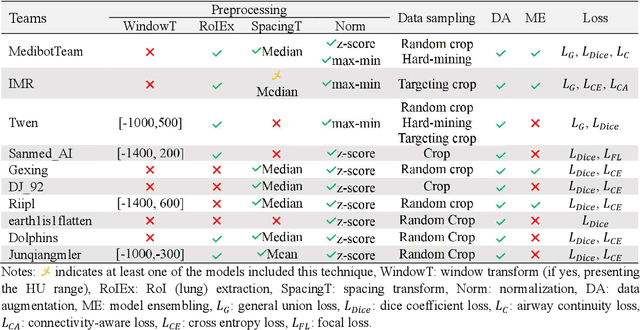
Abstract:Airway-related quantitative imaging biomarkers are crucial for examination, diagnosis, and prognosis in pulmonary diseases. However, the manual delineation of airway trees remains prohibitively time-consuming. While significant efforts have been made towards enhancing airway modelling, current public-available datasets concentrate on lung diseases with moderate morphological variations. The intricate honeycombing patterns present in the lung tissues of fibrotic lung disease patients exacerbate the challenges, often leading to various prediction errors. To address this issue, the 'Airway-Informed Quantitative CT Imaging Biomarker for Fibrotic Lung Disease 2023' (AIIB23) competition was organized in conjunction with the official 2023 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). The airway structures were meticulously annotated by three experienced radiologists. Competitors were encouraged to develop automatic airway segmentation models with high robustness and generalization abilities, followed by exploring the most correlated QIB of mortality prediction. A training set of 120 high-resolution computerised tomography (HRCT) scans were publicly released with expert annotations and mortality status. The online validation set incorporated 52 HRCT scans from patients with fibrotic lung disease and the offline test set included 140 cases from fibrosis and COVID-19 patients. The results have shown that the capacity of extracting airway trees from patients with fibrotic lung disease could be enhanced by introducing voxel-wise weighted general union loss and continuity loss. In addition to the competitive image biomarkers for prognosis, a strong airway-derived biomarker (Hazard ratio>1.5, p<0.0001) was revealed for survival prognostication compared with existing clinical measurements, clinician assessment and AI-based biomarkers.
Modeling 3D cardiac contraction and relaxation with point cloud deformation networks
Jul 20, 2023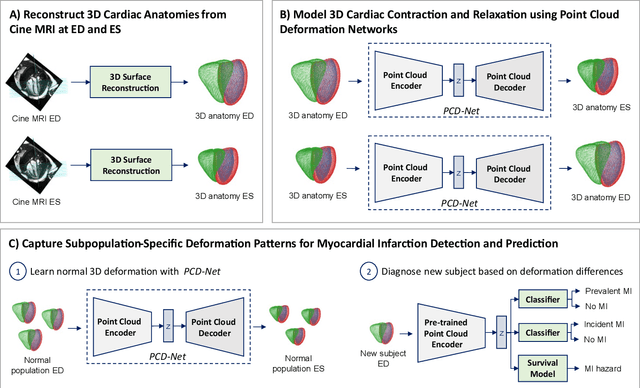
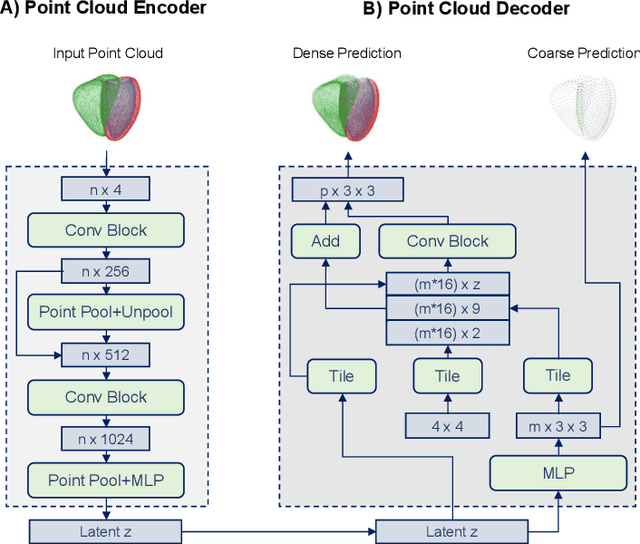
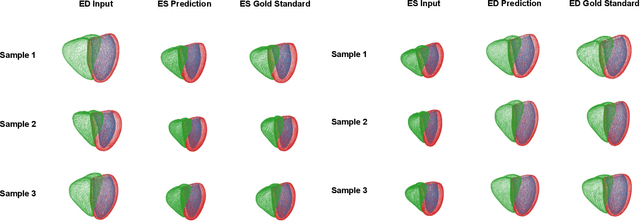
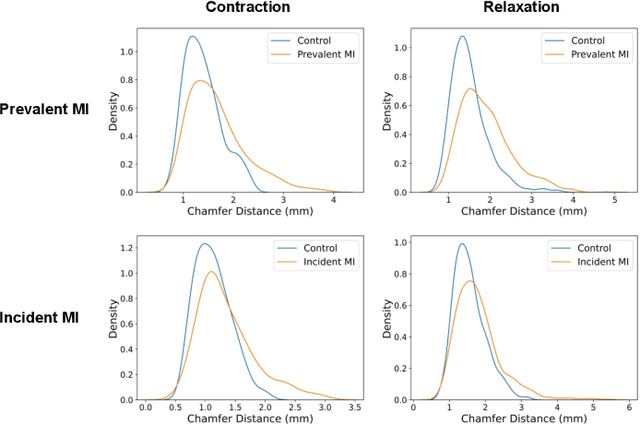
Abstract:Global single-valued biomarkers of cardiac function typically used in clinical practice, such as ejection fraction, provide limited insight on the true 3D cardiac deformation process and hence, limit the understanding of both healthy and pathological cardiac mechanics. In this work, we propose the Point Cloud Deformation Network (PCD-Net) as a novel geometric deep learning approach to model 3D cardiac contraction and relaxation between the extreme ends of the cardiac cycle. It employs the recent advances in point cloud-based deep learning into an encoder-decoder structure, in order to enable efficient multi-scale feature learning directly on multi-class 3D point cloud representations of the cardiac anatomy. We evaluate our approach on a large dataset of over 10,000 cases from the UK Biobank study and find average Chamfer distances between the predicted and ground truth anatomies below the pixel resolution of the underlying image acquisition. Furthermore, we observe similar clinical metrics between predicted and ground truth populations and show that the PCD-Net can successfully capture subpopulation-specific differences between normal subjects and myocardial infarction (MI) patients. We then demonstrate that the learned 3D deformation patterns outperform multiple clinical benchmarks by 13% and 7% in terms of area under the receiver operating characteristic curve for the tasks of prevalent MI detection and incident MI prediction and by 7% in terms of Harrell's concordance index for MI survival analysis.
Multi-objective point cloud autoencoders for explainable myocardial infarction prediction
Jul 20, 2023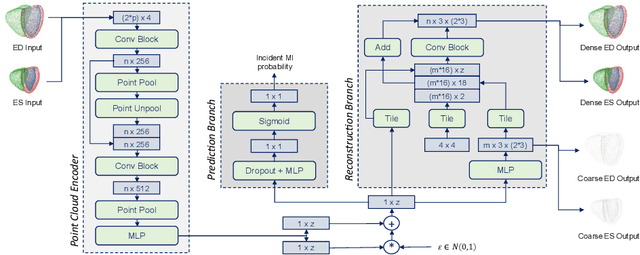
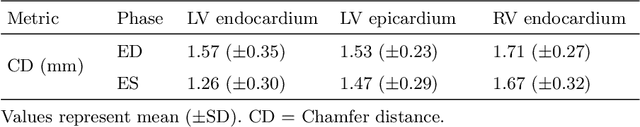
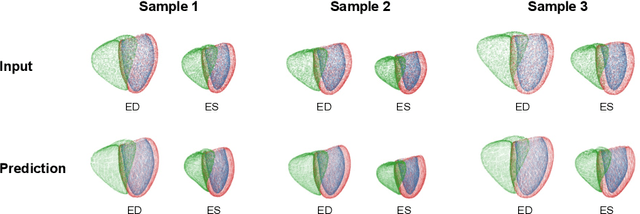
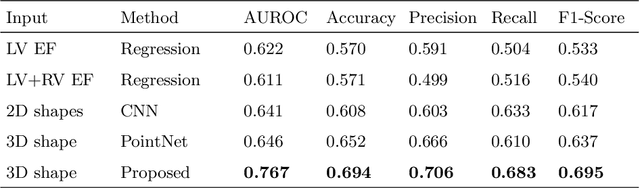
Abstract:Myocardial infarction (MI) is one of the most common causes of death in the world. Image-based biomarkers commonly used in the clinic, such as ejection fraction, fail to capture more complex patterns in the heart's 3D anatomy and thus limit diagnostic accuracy. In this work, we present the multi-objective point cloud autoencoder as a novel geometric deep learning approach for explainable infarction prediction, based on multi-class 3D point cloud representations of cardiac anatomy and function. Its architecture consists of multiple task-specific branches connected by a low-dimensional latent space to allow for effective multi-objective learning of both reconstruction and MI prediction, while capturing pathology-specific 3D shape information in an interpretable latent space. Furthermore, its hierarchical branch design with point cloud-based deep learning operations enables efficient multi-scale feature learning directly on high-resolution anatomy point clouds. In our experiments on a large UK Biobank dataset, the multi-objective point cloud autoencoder is able to accurately reconstruct multi-temporal 3D shapes with Chamfer distances between predicted and input anatomies below the underlying images' pixel resolution. Our method outperforms multiple machine learning and deep learning benchmarks for the task of incident MI prediction by 19% in terms of Area Under the Receiver Operating Characteristic curve. In addition, its task-specific compact latent space exhibits easily separable control and MI clusters with clinically plausible associations between subject encodings and corresponding 3D shapes, thus demonstrating the explainability of the prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge