Hannah Smith
Personalized Topology-Informed 12-Lead ECG Electrode Localization from Incomplete Cardiac MRIs for Efficient Cardiac Digital Twins
Aug 25, 2024Abstract:Cardiac digital twins (CDTs) offer personalized \textit{in-silico} cardiac representations for the inference of multi-scale properties tied to cardiac mechanisms. The creation of CDTs requires precise information about the electrode position on the torso, especially for the personalized electrocardiogram (ECG) calibration. However, current studies commonly rely on additional acquisition of torso imaging and manual/semi-automatic methods for ECG electrode localization. In this study, we propose a novel and efficient topology-informed model to fully automatically extract personalized ECG electrode locations from 2D clinically standard cardiac MRIs. Specifically, we obtain the sparse torso contours from the cardiac MRIs and then localize the electrodes from the contours. Cardiac MRIs aim at imaging of the heart instead of the torso, leading to incomplete torso geometry within the imaging. To tackle the missing topology, we incorporate the electrodes as a subset of the keypoints, which can be explicitly aligned with the 3D torso topology. The experimental results demonstrate that the proposed model outperforms the time-consuming conventional method in terms of accuracy (Euclidean distance: $1.24 \pm 0.293$ cm vs. $1.48 \pm 0.362$ cm) and efficiency ($2$~s vs. $30$-$35$~min). We further demonstrate the effectiveness of using the detected electrodes for \textit{in-silico} ECG simulation, highlighting their potential for creating accurate and efficient CDT models. The code will be released publicly after the manuscript is accepted for publication.
Explaining Answers with Entailment Trees
Apr 17, 2021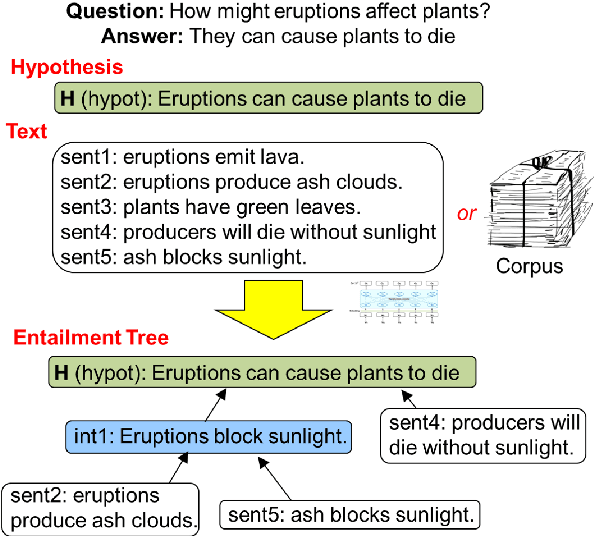
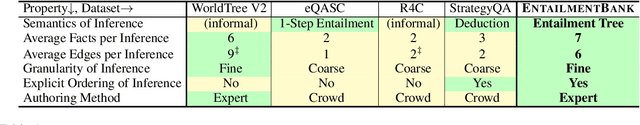
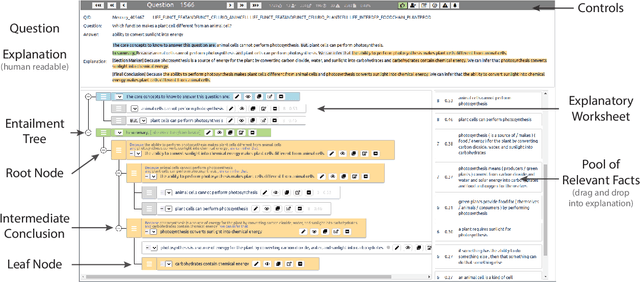

Abstract:Our goal, in the context of open-domain textual question-answering (QA), is to explain answers by not just listing supporting textual evidence ("rationales"), but also showing how such evidence leads to the answer in a systematic way. If this could be done, new opportunities for understanding and debugging the system's reasoning would become possible. Our approach is to generate explanations in the form of entailment trees, namely a tree of entailment steps from facts that are known, through intermediate conclusions, to the final answer. To train a model with this skill, we created ENTAILMENTBANK, the first dataset to contain multistep entailment trees. At each node in the tree (typically) two or more facts compose together to produce a new conclusion. Given a hypothesis (question + answer), we define three increasingly difficult explanation tasks: generate a valid entailment tree given (a) all relevant sentences (the leaves of the gold entailment tree), (b) all relevant and some irrelevant sentences, or (c) a corpus. We show that a strong language model only partially solves these tasks, and identify several new directions to improve performance. This work is significant as it provides a new type of dataset (multistep entailments) and baselines, offering a new avenue for the community to generate richer, more systematic explanations.
ScienceExamCER: A High-Density Fine-Grained Science-Domain Corpus for Common Entity Recognition
Nov 24, 2019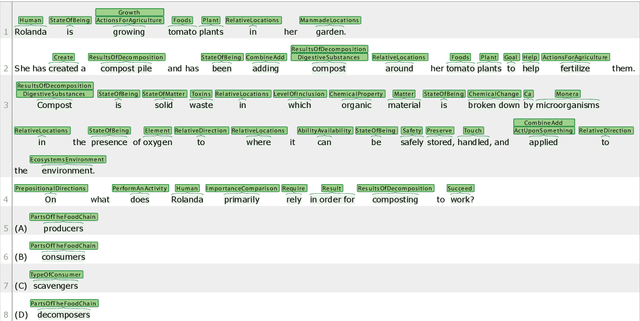
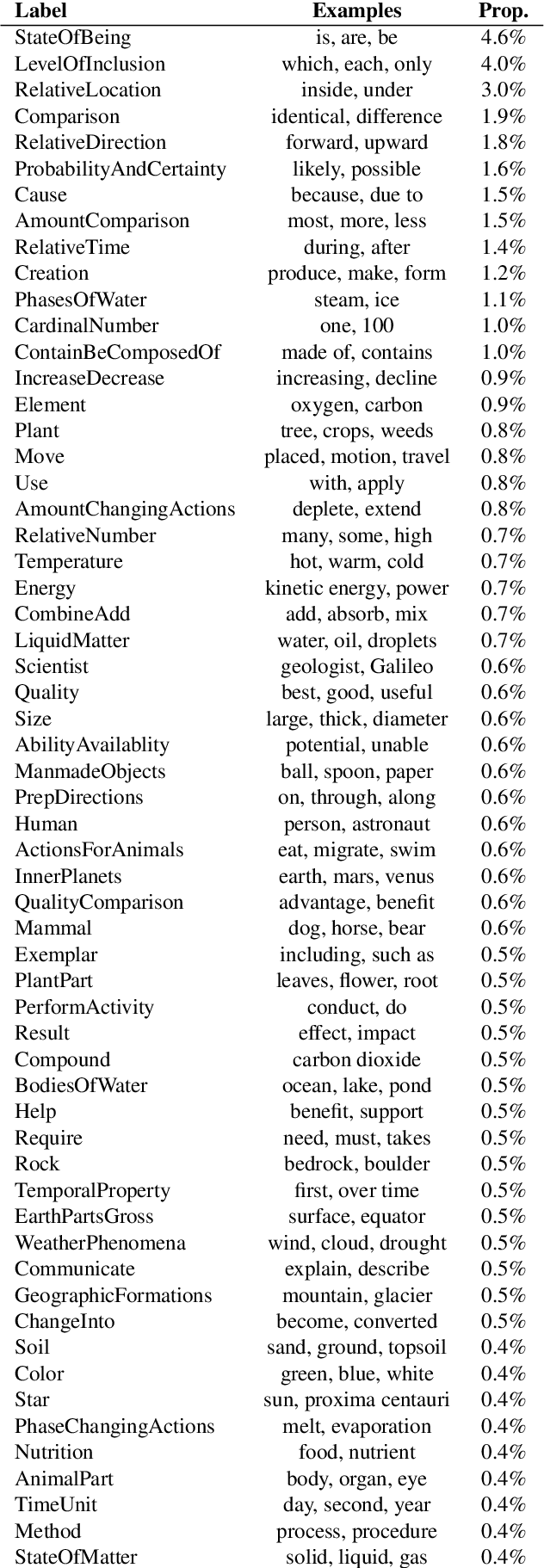
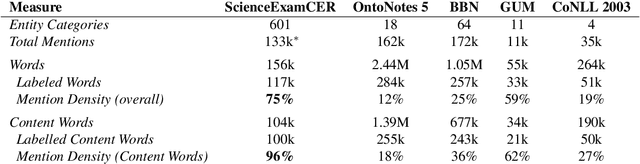
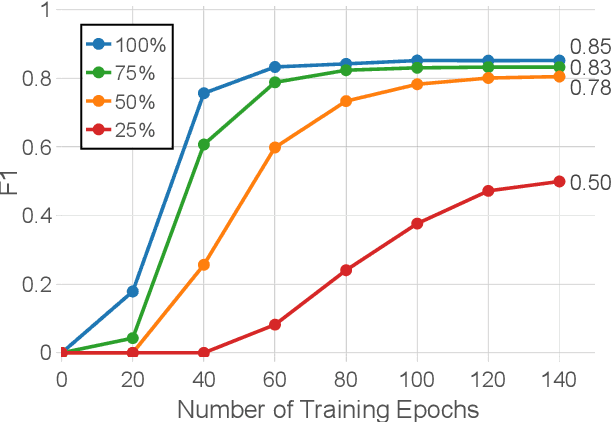
Abstract:Named entity recognition identifies common classes of entities in text, but these entity labels are generally sparse, limiting utility to downstream tasks. In this work we present ScienceExamCER, a densely-labeled semantic classification corpus of 133k mentions in the science exam domain where nearly all (96%) of content words have been annotated with one or more fine-grained semantic class labels including taxonomic groups, meronym groups, verb/action groups, properties and values, and synonyms. Semantic class labels are drawn from a manually-constructed fine-grained typology of 601 classes generated through a data-driven analysis of 4,239 science exam questions. We show an off-the-shelf BERT-based named entity recognition model modified for multi-label classification achieves an accuracy of 0.85 F1 on this task, suggesting strong utility for downstream tasks in science domain question answering requiring densely-labeled semantic classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge