Julia Camps
Jenny
Personalized Topology-Informed 12-Lead ECG Electrode Localization from Incomplete Cardiac MRIs for Efficient Cardiac Digital Twins
Aug 25, 2024Abstract:Cardiac digital twins (CDTs) offer personalized \textit{in-silico} cardiac representations for the inference of multi-scale properties tied to cardiac mechanisms. The creation of CDTs requires precise information about the electrode position on the torso, especially for the personalized electrocardiogram (ECG) calibration. However, current studies commonly rely on additional acquisition of torso imaging and manual/semi-automatic methods for ECG electrode localization. In this study, we propose a novel and efficient topology-informed model to fully automatically extract personalized ECG electrode locations from 2D clinically standard cardiac MRIs. Specifically, we obtain the sparse torso contours from the cardiac MRIs and then localize the electrodes from the contours. Cardiac MRIs aim at imaging of the heart instead of the torso, leading to incomplete torso geometry within the imaging. To tackle the missing topology, we incorporate the electrodes as a subset of the keypoints, which can be explicitly aligned with the 3D torso topology. The experimental results demonstrate that the proposed model outperforms the time-consuming conventional method in terms of accuracy (Euclidean distance: $1.24 \pm 0.293$ cm vs. $1.48 \pm 0.362$ cm) and efficiency ($2$~s vs. $30$-$35$~min). We further demonstrate the effectiveness of using the detected electrodes for \textit{in-silico} ECG simulation, highlighting their potential for creating accurate and efficient CDT models. The code will be released publicly after the manuscript is accepted for publication.
Solving the Inverse Problem of Electrocardiography for Cardiac Digital Twins: A Survey
Jun 17, 2024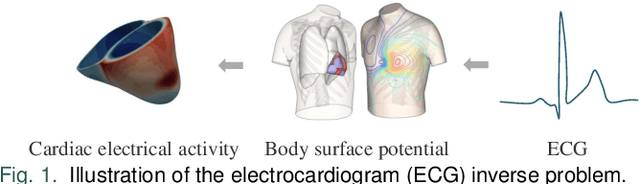
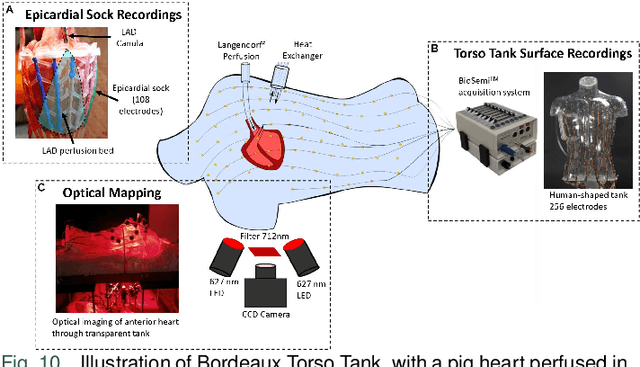
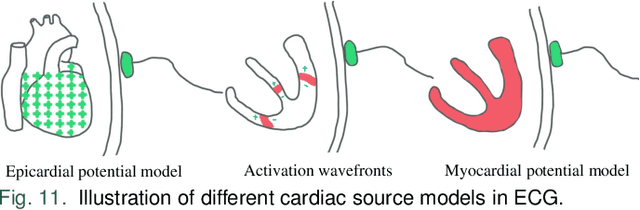
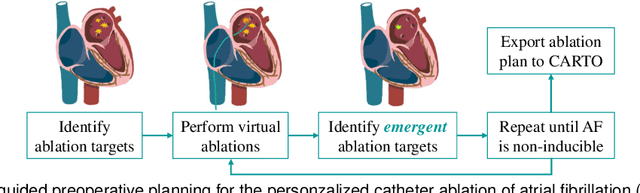
Abstract:Cardiac digital twins are personalized virtual representations used to understand complex heart mechanisms. Solving the ECG inverse problem is crucial for accurate virtual heart modelling, enabling the derivation of internal electrical activity information from recorded surface potentials. Despite challenges from cardiac complexity, noisy ECG data, and computational efficiency, recent advancements hold significant promise for enhancing virtual heart modelling, ultimately advancing precision medicine in cardiology. This paper aims to provide a comprehensive review of the methods of solving ECG inverse problem, the validation strategies, the clinical applications, and future perspectives. For the computing methodologies, we broadly classify state-of-the-art approaches into two categories: deterministic and probabilistic methods, including conventional and deep learning-based techniques. Integrating physics laws with deep learning models holds promise, but challenges such as capturing dynamic electrophysiology accurately, accessing accurate domain knowledge, and quantifying prediction uncertainty persist. Integrating models into clinical workflows while ensuring interpretability and usability for healthcare professionals is essential. Overcoming these challenges will drive further research in cardiac digital twins.
Towards Enabling Cardiac Digital Twins of Myocardial Infarction Using Deep Computational Models for Inverse Inference
Jul 13, 2023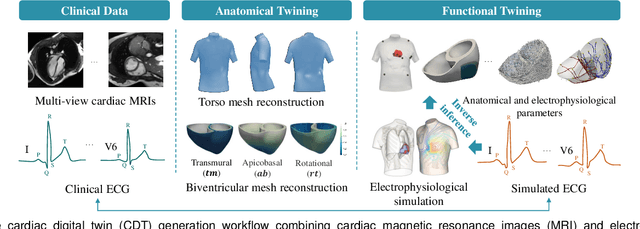
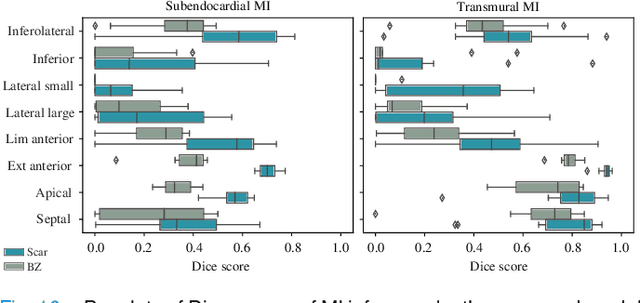
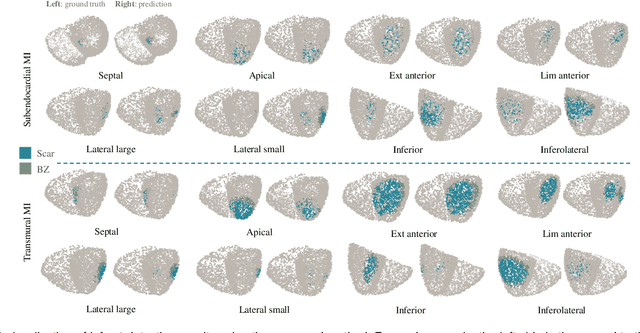
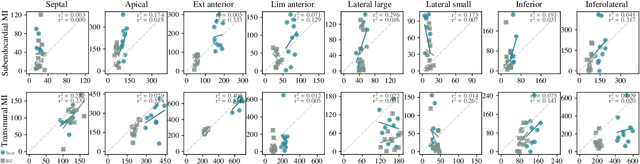
Abstract:Myocardial infarction (MI) demands precise and swift diagnosis. Cardiac digital twins (CDTs) have the potential to offer individualized evaluation of cardiac function in a non-invasive manner, making them a promising approach for personalized diagnosis and treatment planning of MI. The inference of accurate myocardial tissue properties is crucial in creating a reliable CDT platform, and particularly in the context of studying MI. In this work, we investigate the feasibility of inferring myocardial tissue properties from the electrocardiogram (ECG), focusing on the development of a comprehensive CDT platform specifically designed for MI. The platform integrates multi-modal data, such as cardiac MRI and ECG, to enhance the accuracy and reliability of the inferred tissue properties. We perform a sensitivity analysis based on computer simulations, systematically exploring the effects of infarct location, size, degree of transmurality, and electrical activity alteration on the simulated QRS complex of ECG, to establish the limits of the approach. We subsequently propose a deep computational model to infer infarct location and distribution from the simulated QRS. The in silico experimental results show that our model can effectively capture the complex relationships between the QRS signals and the corresponding infarct regions, with promising potential for clinical application in the future. The code will be released publicly once the manuscript is accepted for publication.
Influence of Myocardial Infarction on QRS Properties: A Simulation Study
Apr 21, 2023



Abstract:The interplay between structural and electrical changes in the heart after myocardial infarction (MI) plays a key role in the initiation and maintenance of arrhythmia. The anatomical and electrophysiological properties of scar, border zone, and normal myocardium modify the electrocardiographic morphology, which is routinely analysed in clinical settings. However, the influence of various MI properties on the QRS is not intuitively predictable.In this work, we have systematically investigated the effects of 17 post-MI scenarios, varying the location, size, transmural extent, and conductive level of scarring and border zone area, on the forward-calculated QRS. Additionally, we have compared the contributions of different QRS score criteria for quantifying post-MI pathophysiology.The propagation of electrical activity in the ventricles is simulated via a Eikonal model on a unified coordinate system.The analysis has been performed on 49 subjects, and the results imply that the QRS is capable of identifying MI, suggesting the feasibility of inversely reconstructing infarct regions from QRS.There exist sensitivity variations of different QRS criteria for identifying 17 MI scenarios, which is informative for solving the inverse problem.
Deep Computational Model for the Inference of Ventricular Activation Properties
Aug 08, 2022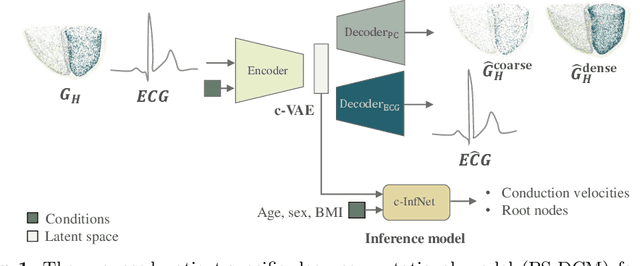
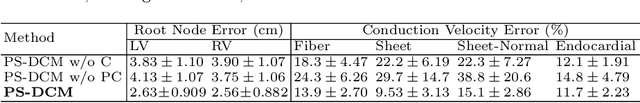
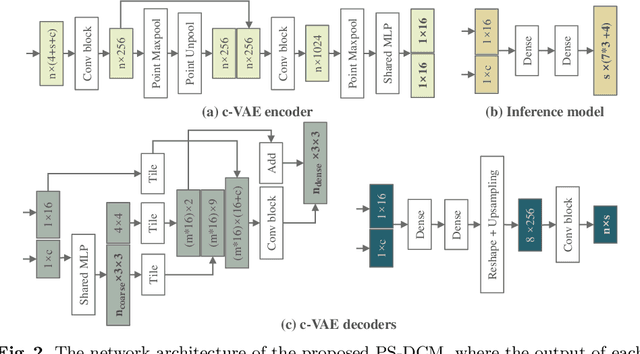

Abstract:Patient-specific cardiac computational models are essential for the efficient realization of precision medicine and in-silico clinical trials using digital twins. Cardiac digital twins can provide non-invasive characterizations of cardiac functions for individual patients, and therefore are promising for the patient-specific diagnosis and therapy stratification. However, current workflows for both the anatomical and functional twinning phases, referring to the inference of model anatomy and parameter from clinical data, are not sufficiently efficient, robust, and accurate. In this work, we propose a deep learning based patient-specific computational model, which can fuse both anatomical and electrophysiological information for the inference of ventricular activation properties, i.e., conduction velocities and root nodes. The activation properties can provide a quantitative assessment of cardiac electrophysiological function for the guidance of interventional procedures. We employ the Eikonal model to generate simulated electrocardiogram (ECG) with ground truth properties to train the inference model, where specific patient information has also been considered. For evaluation, we test the model on the simulated data and obtain generally promising results with fast computational time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge