Marcel Beetz
Large Language Model-informed ECG Dual Attention Network for Heart Failure Risk Prediction
Mar 22, 2024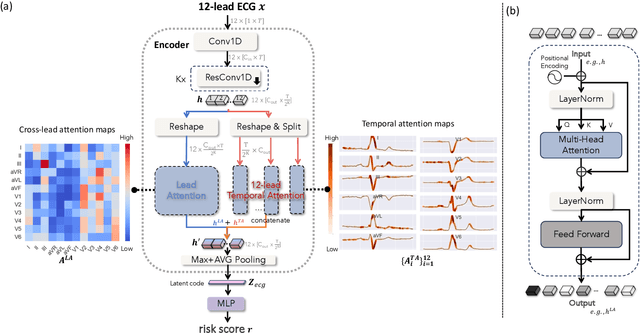
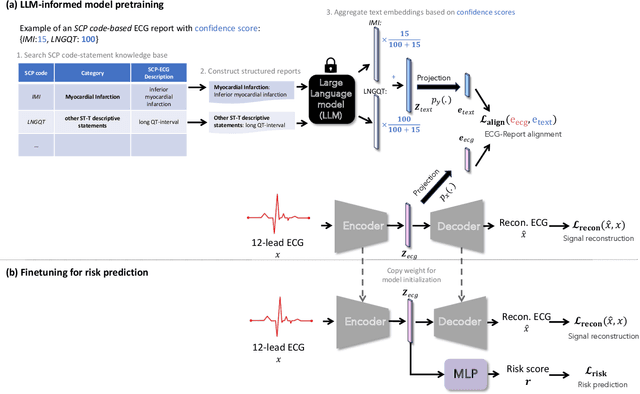
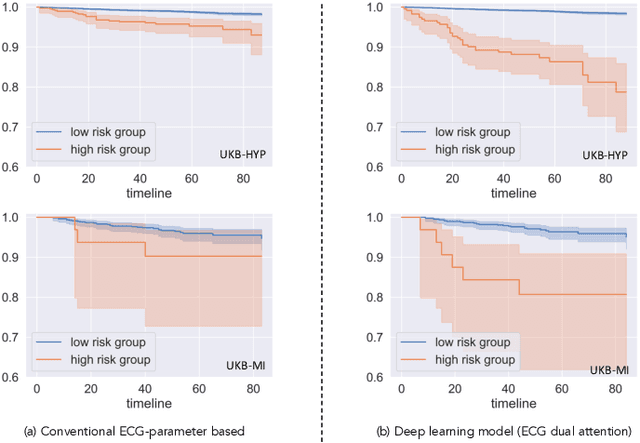
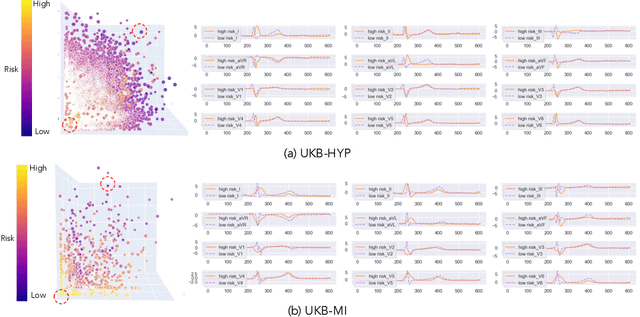
Abstract:Heart failure (HF) poses a significant public health challenge, with a rising global mortality rate. Early detection and prevention of HF could significantly reduce its impact. We introduce a novel methodology for predicting HF risk using 12-lead electrocardiograms (ECGs). We present a novel, lightweight dual-attention ECG network designed to capture complex ECG features essential for early HF risk prediction, despite the notable imbalance between low and high-risk groups. This network incorporates a cross-lead attention module and twelve lead-specific temporal attention modules, focusing on cross-lead interactions and each lead's local dynamics. To further alleviate model overfitting, we leverage a large language model (LLM) with a public ECG-Report dataset for pretraining on an ECG-report alignment task. The network is then fine-tuned for HF risk prediction using two specific cohorts from the UK Biobank study, focusing on patients with hypertension (UKB-HYP) and those who have had a myocardial infarction (UKB-MI).The results reveal that LLM-informed pre-training substantially enhances HF risk prediction in these cohorts. The dual-attention design not only improves interpretability but also predictive accuracy, outperforming existing competitive methods with C-index scores of 0.6349 for UKB-HYP and 0.5805 for UKB-MI. This demonstrates our method's potential in advancing HF risk assessment with clinical complex ECG data.
Anatomical basis of sex differences in human post-myocardial infarction ECG phenotypes identified by novel automated torso-cardiac 3D reconstruction
Dec 21, 2023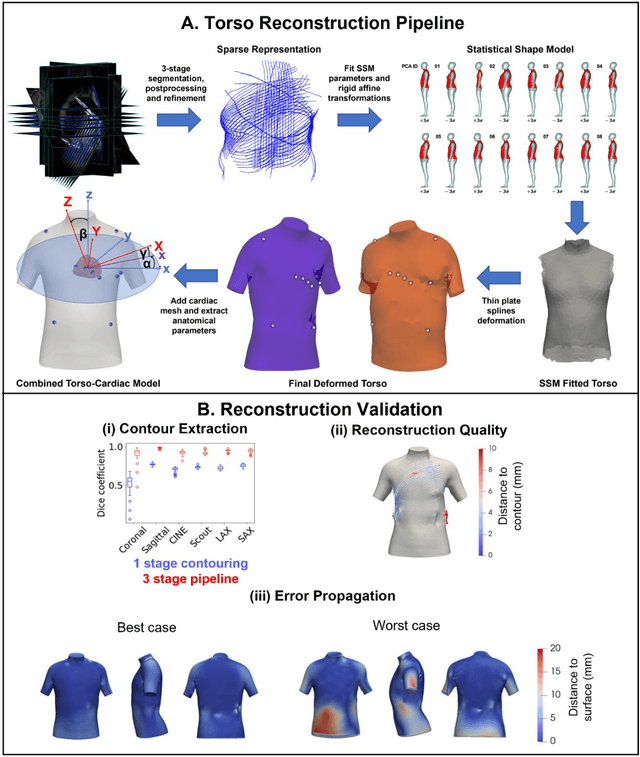
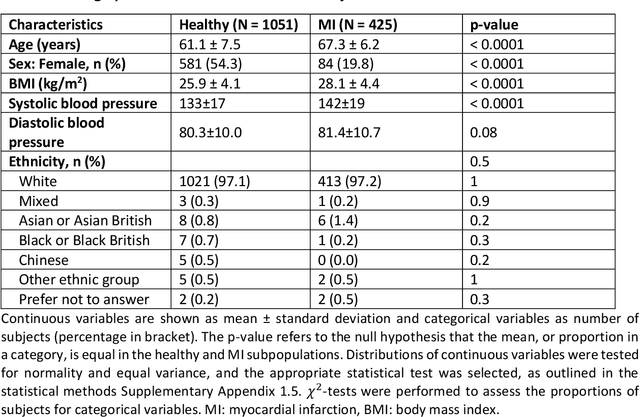
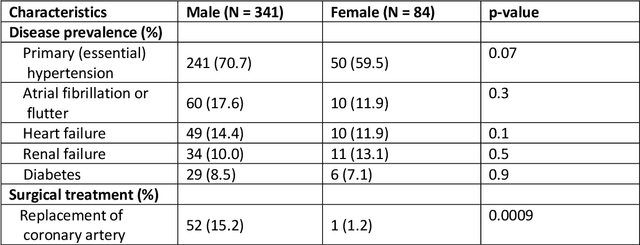
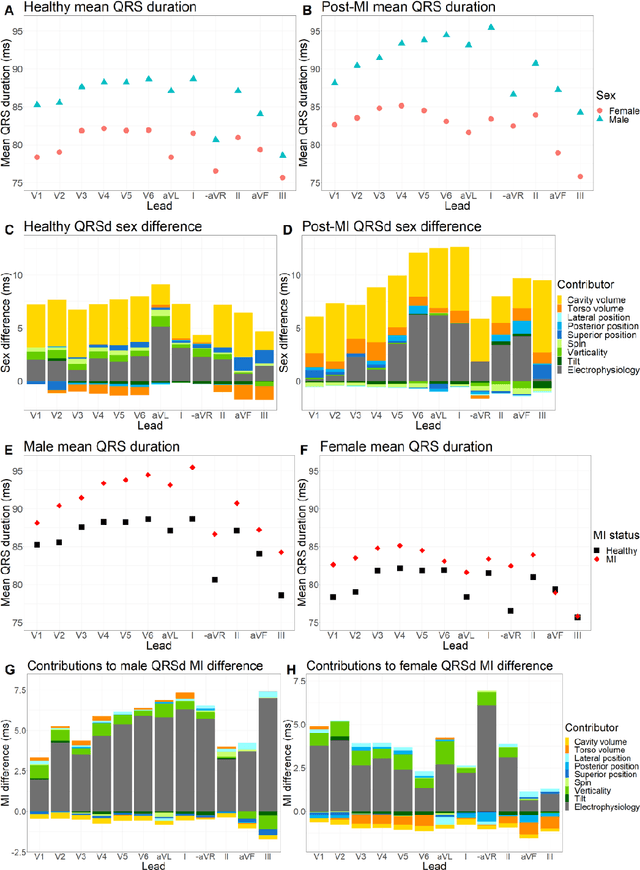
Abstract:The electrocardiogram (ECG) is routinely used in cardiology, though its interpretation is confounded by anatomical variability. A novel, automated computational pipeline enables quantification of torso-ventricular anatomy metrics from magnetic resonance imaging, and comparison to ECG characteristics. Sex and myocardial infarction differences are investigated based on 1051 healthy and 425 post-MI subjects from UK Biobank. Smaller ventricles in females explain ~50% of shorter QRS durations than in males, and contribute to lower STJ amplitudes in females (also due to more superior and posterior position). In females, torso-ventricular anatomy, particularly from larger BMI, is a stronger modulator of T wave amplitude reductions and left-deviated R axis angles in post-MI than in males. Thus, female MI phenotype is less reflective of pathology, and baseline STJ amplitudes and QRS durations are further from clinical thresholds. Therefore, quantification of anatomical sex-differences and impact on ECG in health and disease is critical to avoid clinical sex-bias.
Multi-objective point cloud autoencoders for explainable myocardial infarction prediction
Jul 20, 2023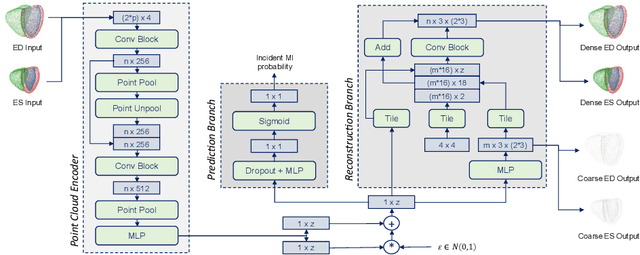
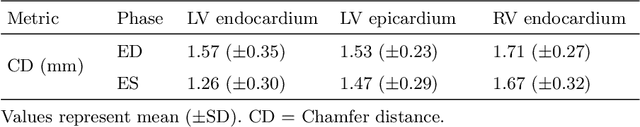
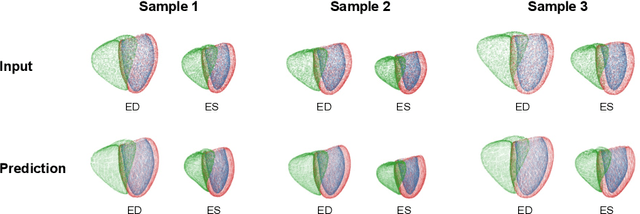
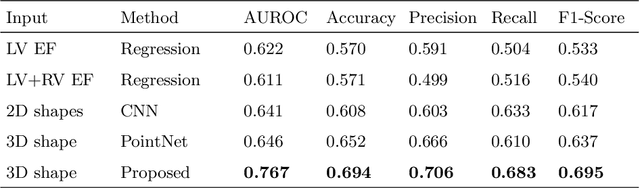
Abstract:Myocardial infarction (MI) is one of the most common causes of death in the world. Image-based biomarkers commonly used in the clinic, such as ejection fraction, fail to capture more complex patterns in the heart's 3D anatomy and thus limit diagnostic accuracy. In this work, we present the multi-objective point cloud autoencoder as a novel geometric deep learning approach for explainable infarction prediction, based on multi-class 3D point cloud representations of cardiac anatomy and function. Its architecture consists of multiple task-specific branches connected by a low-dimensional latent space to allow for effective multi-objective learning of both reconstruction and MI prediction, while capturing pathology-specific 3D shape information in an interpretable latent space. Furthermore, its hierarchical branch design with point cloud-based deep learning operations enables efficient multi-scale feature learning directly on high-resolution anatomy point clouds. In our experiments on a large UK Biobank dataset, the multi-objective point cloud autoencoder is able to accurately reconstruct multi-temporal 3D shapes with Chamfer distances between predicted and input anatomies below the underlying images' pixel resolution. Our method outperforms multiple machine learning and deep learning benchmarks for the task of incident MI prediction by 19% in terms of Area Under the Receiver Operating Characteristic curve. In addition, its task-specific compact latent space exhibits easily separable control and MI clusters with clinically plausible associations between subject encodings and corresponding 3D shapes, thus demonstrating the explainability of the prediction.
Modeling 3D cardiac contraction and relaxation with point cloud deformation networks
Jul 20, 2023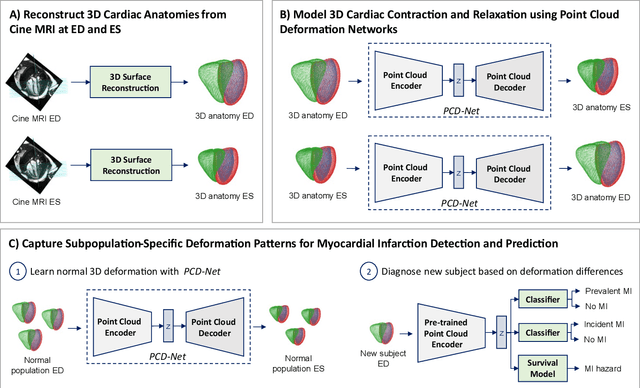
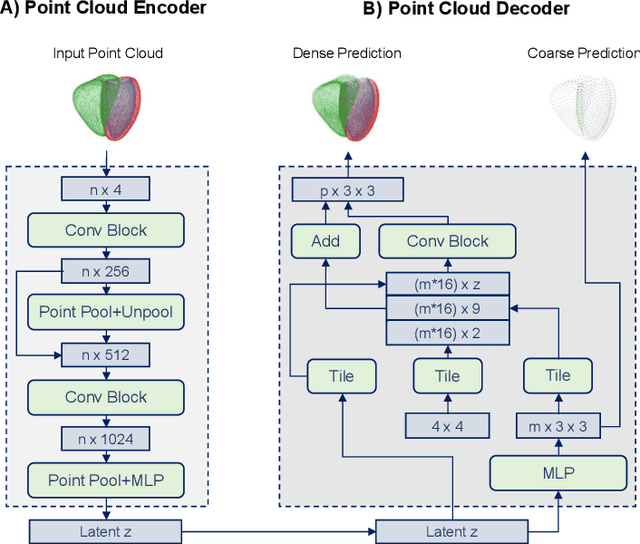
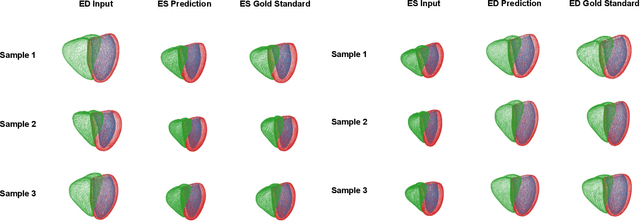
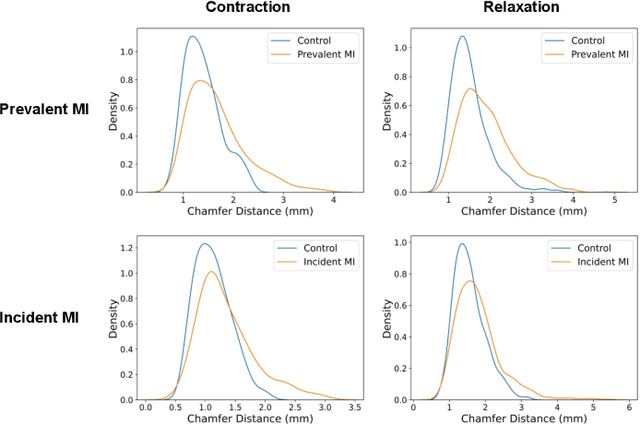
Abstract:Global single-valued biomarkers of cardiac function typically used in clinical practice, such as ejection fraction, provide limited insight on the true 3D cardiac deformation process and hence, limit the understanding of both healthy and pathological cardiac mechanics. In this work, we propose the Point Cloud Deformation Network (PCD-Net) as a novel geometric deep learning approach to model 3D cardiac contraction and relaxation between the extreme ends of the cardiac cycle. It employs the recent advances in point cloud-based deep learning into an encoder-decoder structure, in order to enable efficient multi-scale feature learning directly on multi-class 3D point cloud representations of the cardiac anatomy. We evaluate our approach on a large dataset of over 10,000 cases from the UK Biobank study and find average Chamfer distances between the predicted and ground truth anatomies below the pixel resolution of the underlying image acquisition. Furthermore, we observe similar clinical metrics between predicted and ground truth populations and show that the PCD-Net can successfully capture subpopulation-specific differences between normal subjects and myocardial infarction (MI) patients. We then demonstrate that the learned 3D deformation patterns outperform multiple clinical benchmarks by 13% and 7% in terms of area under the receiver operating characteristic curve for the tasks of prevalent MI detection and incident MI prediction and by 7% in terms of Harrell's concordance index for MI survival analysis.
Multi-class point cloud completion networks for 3D cardiac anatomy reconstruction from cine magnetic resonance images
Jul 18, 2023Abstract:Cine magnetic resonance imaging (MRI) is the current gold standard for the assessment of cardiac anatomy and function. However, it typically only acquires a set of two-dimensional (2D) slices of the underlying three-dimensional (3D) anatomy of the heart, thus limiting the understanding and analysis of both healthy and pathological cardiac morphology and physiology. In this paper, we propose a novel fully automatic surface reconstruction pipeline capable of reconstructing multi-class 3D cardiac anatomy meshes from raw cine MRI acquisitions. Its key component is a multi-class point cloud completion network (PCCN) capable of correcting both the sparsity and misalignment issues of the 3D reconstruction task in a unified model. We first evaluate the PCCN on a large synthetic dataset of biventricular anatomies and observe Chamfer distances between reconstructed and gold standard anatomies below or similar to the underlying image resolution for multiple levels of slice misalignment. Furthermore, we find a reduction in reconstruction error compared to a benchmark 3D U-Net by 32% and 24% in terms of Hausdorff distance and mean surface distance, respectively. We then apply the PCCN as part of our automated reconstruction pipeline to 1000 subjects from the UK Biobank study in a cross-domain transfer setting and demonstrate its ability to reconstruct accurate and topologically plausible biventricular heart meshes with clinical metrics comparable to the previous literature. Finally, we investigate the robustness of our proposed approach and observe its capacity to successfully handle multiple common outlier conditions.
3D Shape-Based Myocardial Infarction Prediction Using Point Cloud Classification Networks
Jul 14, 2023



Abstract:Myocardial infarction (MI) is one of the most prevalent cardiovascular diseases with associated clinical decision-making typically based on single-valued imaging biomarkers. However, such metrics only approximate the complex 3D structure and physiology of the heart and hence hinder a better understanding and prediction of MI outcomes. In this work, we investigate the utility of complete 3D cardiac shapes in the form of point clouds for an improved detection of MI events. To this end, we propose a fully automatic multi-step pipeline consisting of a 3D cardiac surface reconstruction step followed by a point cloud classification network. Our method utilizes recent advances in geometric deep learning on point clouds to enable direct and efficient multi-scale learning on high-resolution surface models of the cardiac anatomy. We evaluate our approach on 1068 UK Biobank subjects for the tasks of prevalent MI detection and incident MI prediction and find improvements of ~13% and ~5% respectively over clinical benchmarks. Furthermore, we analyze the role of each ventricle and cardiac phase for 3D shape-based MI detection and conduct a visual analysis of the morphological and physiological patterns typically associated with MI outcomes.
Towards Enabling Cardiac Digital Twins of Myocardial Infarction Using Deep Computational Models for Inverse Inference
Jul 13, 2023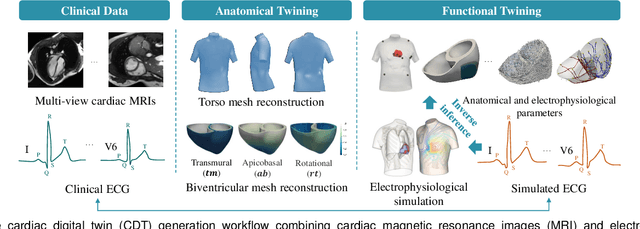
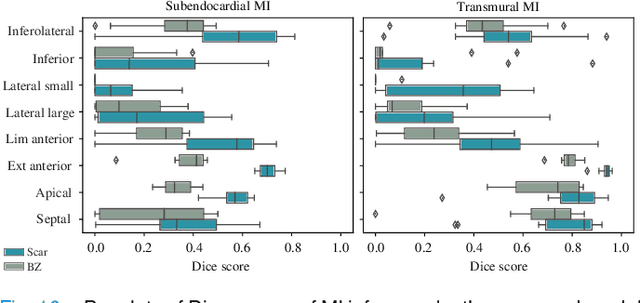
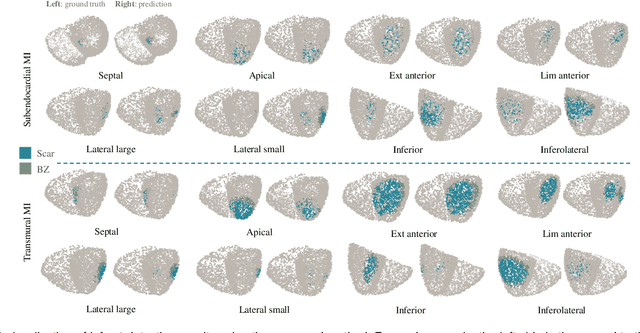
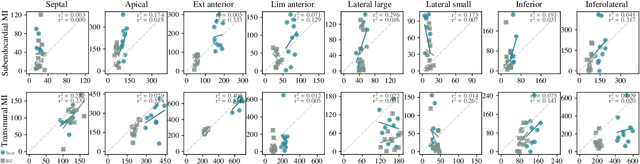
Abstract:Myocardial infarction (MI) demands precise and swift diagnosis. Cardiac digital twins (CDTs) have the potential to offer individualized evaluation of cardiac function in a non-invasive manner, making them a promising approach for personalized diagnosis and treatment planning of MI. The inference of accurate myocardial tissue properties is crucial in creating a reliable CDT platform, and particularly in the context of studying MI. In this work, we investigate the feasibility of inferring myocardial tissue properties from the electrocardiogram (ECG), focusing on the development of a comprehensive CDT platform specifically designed for MI. The platform integrates multi-modal data, such as cardiac MRI and ECG, to enhance the accuracy and reliability of the inferred tissue properties. We perform a sensitivity analysis based on computer simulations, systematically exploring the effects of infarct location, size, degree of transmurality, and electrical activity alteration on the simulated QRS complex of ECG, to establish the limits of the approach. We subsequently propose a deep computational model to infer infarct location and distribution from the simulated QRS. The in silico experimental results show that our model can effectively capture the complex relationships between the QRS signals and the corresponding infarct regions, with promising potential for clinical application in the future. The code will be released publicly once the manuscript is accepted for publication.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Deep Computational Model for the Inference of Ventricular Activation Properties
Aug 08, 2022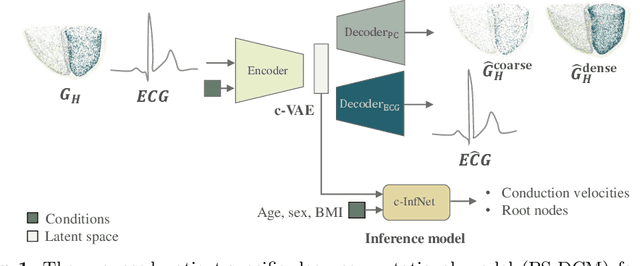
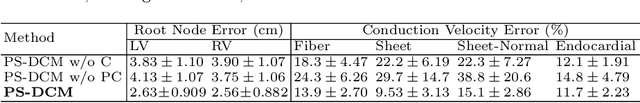
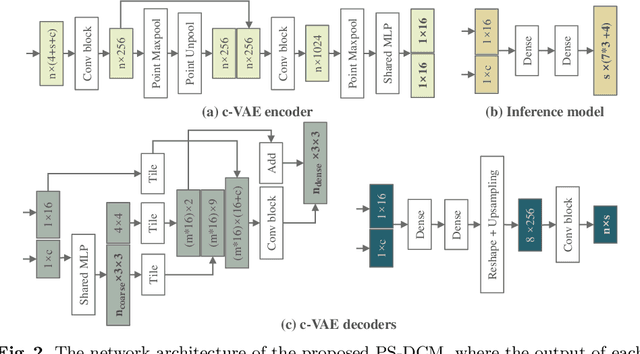

Abstract:Patient-specific cardiac computational models are essential for the efficient realization of precision medicine and in-silico clinical trials using digital twins. Cardiac digital twins can provide non-invasive characterizations of cardiac functions for individual patients, and therefore are promising for the patient-specific diagnosis and therapy stratification. However, current workflows for both the anatomical and functional twinning phases, referring to the inference of model anatomy and parameter from clinical data, are not sufficiently efficient, robust, and accurate. In this work, we propose a deep learning based patient-specific computational model, which can fuse both anatomical and electrophysiological information for the inference of ventricular activation properties, i.e., conduction velocities and root nodes. The activation properties can provide a quantitative assessment of cardiac electrophysiological function for the guidance of interventional procedures. We employ the Eikonal model to generate simulated electrocardiogram (ECG) with ground truth properties to train the inference model, where specific patient information has also been considered. For evaluation, we test the model on the simulated data and obtain generally promising results with fast computational time.
The Liver Tumor Segmentation Benchmark (LiTS)
Jan 13, 2019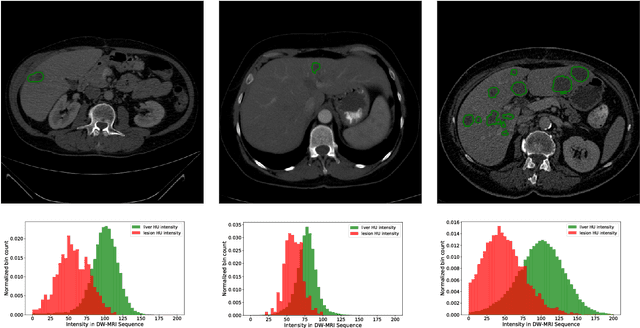
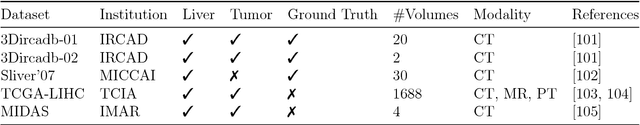
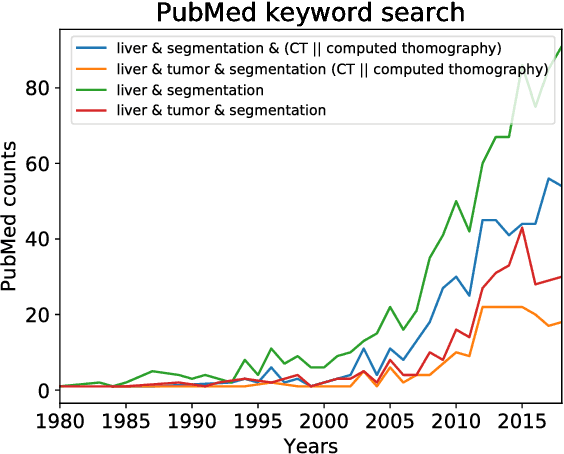
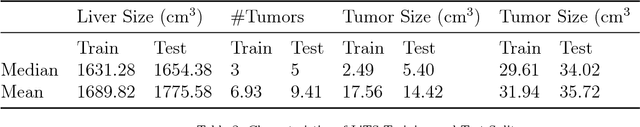
Abstract:In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LITS) organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2016 and International Conference On Medical Image Computing Computer Assisted Intervention (MICCAI) 2017. Twenty four valid state-of-the-art liver and liver tumor segmentation algorithms were applied to a set of 131 computed tomography (CT) volumes with different types of tumor contrast levels (hyper-/hypo-intense), abnormalities in tissues (metastasectomie) size and varying amount of lesions. The submitted algorithms have been tested on 70 undisclosed volumes. The dataset is created in collaboration with seven hospitals and research institutions and manually reviewed by independent three radiologists. We found that not a single algorithm performed best for liver and tumors. The best liver segmentation algorithm achieved a Dice score of 0.96(MICCAI) whereas for tumor segmentation the best algorithm evaluated at 0.67(ISBI) and 0.70(MICCAI). The LITS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge