Sharib Ali
NeeCo: Image Synthesis of Novel Instrument States Based on Dynamic and Deformable 3D Gaussian Reconstruction
Aug 11, 2025
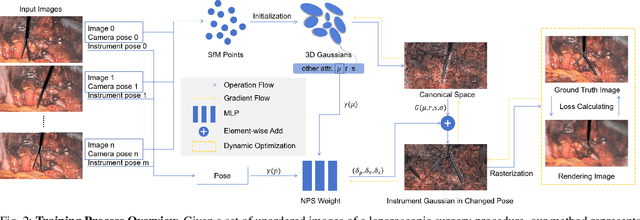

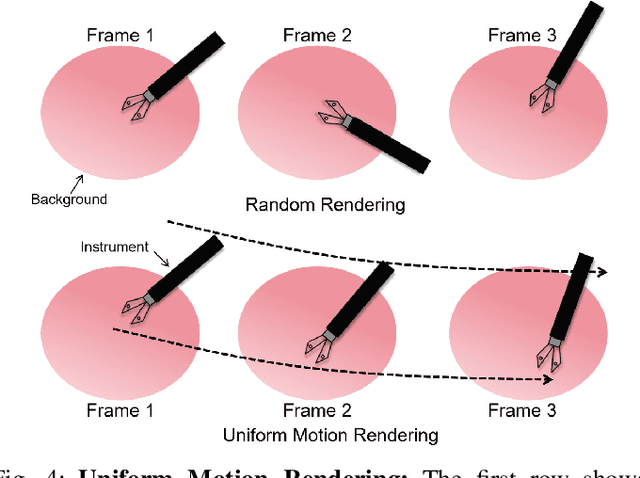
Abstract:Computer vision-based technologies significantly enhance surgical automation by advancing tool tracking, detection, and localization. However, Current data-driven approaches are data-voracious, requiring large, high-quality labeled image datasets, which limits their application in surgical data science. Our Work introduces a novel dynamic Gaussian Splatting technique to address the data scarcity in surgical image datasets. We propose a dynamic Gaussian model to represent dynamic surgical scenes, enabling the rendering of surgical instruments from unseen viewpoints and deformations with real tissue backgrounds. We utilize a dynamic training adjustment strategy to address challenges posed by poorly calibrated camera poses from real-world scenarios. Additionally, we propose a method based on dynamic Gaussians for automatically generating annotations for our synthetic data. For evaluation, we constructed a new dataset featuring seven scenes with 14,000 frames of tool and camera motion and tool jaw articulation, with a background of an ex-vivo porcine model. Using this dataset, we synthetically replicate the scene deformation from the ground truth data, allowing direct comparisons of synthetic image quality. Experimental results illustrate that our method generates photo-realistic labeled image datasets with the highest values in Peak-Signal-to-Noise Ratio (29.87). We further evaluate the performance of medical-specific neural networks trained on real and synthetic images using an unseen real-world image dataset. Our results show that the performance of models trained on synthetic images generated by the proposed method outperforms those trained with state-of-the-art standard data augmentation by 10%, leading to an overall improvement in model performances by nearly 15%.
A Self-Supervised Framework for Improved Generalisability in Ultrasound B-mode Image Segmentation
Feb 04, 2025Abstract:Ultrasound (US) imaging is clinically invaluable due to its noninvasive and safe nature. However, interpreting US images is challenging, requires significant expertise, and time, and is often prone to errors. Deep learning offers assistive solutions such as segmentation. Supervised methods rely on large, high-quality, and consistently labeled datasets, which are challenging to curate. Moreover, these methods tend to underperform on out-of-distribution data, limiting their clinical utility. Self-supervised learning (SSL) has emerged as a promising alternative, leveraging unlabeled data to enhance model performance and generalisability. We introduce a contrastive SSL approach tailored for B-mode US images, incorporating a novel Relation Contrastive Loss (RCL). RCL encourages learning of distinct features by differentiating positive and negative sample pairs through a learnable metric. Additionally, we propose spatial and frequency-based augmentation strategies for the representation learning on US images. Our approach significantly outperforms traditional supervised segmentation methods across three public breast US datasets, particularly in data-limited scenarios. Notable improvements on the Dice similarity metric include a 4% increase on 20% and 50% of the BUSI dataset, nearly 6% and 9% improvements on 20% and 50% of the BrEaST dataset, and 6.4% and 3.7% improvements on 20% and 50% of the UDIAT dataset, respectively. Furthermore, we demonstrate superior generalisability on the out-of-distribution UDIAT dataset with performance boosts of 20.6% and 13.6% compared to the supervised baseline using 20% and 50% of the BUSI and BrEaST training data, respectively. Our research highlights that domain-inspired SSL can improve US segmentation, especially under data-limited conditions.
Solving the long-tailed distribution problem by exploiting the synergies and balance of different techniques
Jan 23, 2025



Abstract:In real-world data, long-tailed data distribution is common, making it challenging for models trained on empirical risk minimisation to learn and classify tail classes effectively. While many studies have sought to improve long tail recognition by altering the data distribution in the feature space and adjusting model decision boundaries, research on the synergy and corrective approach among various methods is limited. Our study delves into three long-tail recognition techniques: Supervised Contrastive Learning (SCL), Rare-Class Sample Generator (RSG), and Label-Distribution-Aware Margin Loss (LDAM). SCL enhances intra-class clusters based on feature similarity and promotes clear inter-class separability but tends to favour dominant classes only. When RSG is integrated into the model, we observed that the intra-class features further cluster towards the class centre, which demonstrates a synergistic effect together with SCL's principle of enhancing intra-class clustering. RSG generates new tail features and compensates for the tail feature space squeezed by SCL. Similarly, LDAM is known to introduce a larger margin specifically for tail classes; we demonstrate that LDAM further bolsters the model's performance on tail classes when combined with the more explicit decision boundaries achieved by SCL and RSG. Furthermore, SCL can compensate for the dominant class accuracy sacrificed by RSG and LDAM. Our research emphasises the synergy and balance among the three techniques, with each amplifying the strengths of the others and mitigating their shortcomings. Our experiment on long-tailed distribution datasets, using an end-to-end architecture, yields competitive results by enhancing tail class accuracy without compromising dominant class performance, achieving a balanced improvement across all classes.
NCDD: Nearest Centroid Distance Deficit for Out-Of-Distribution Detection in Gastrointestinal Vision
Dec 02, 2024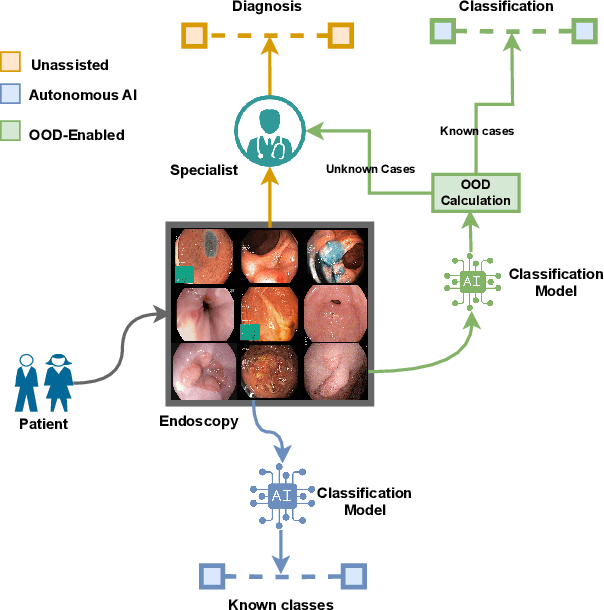

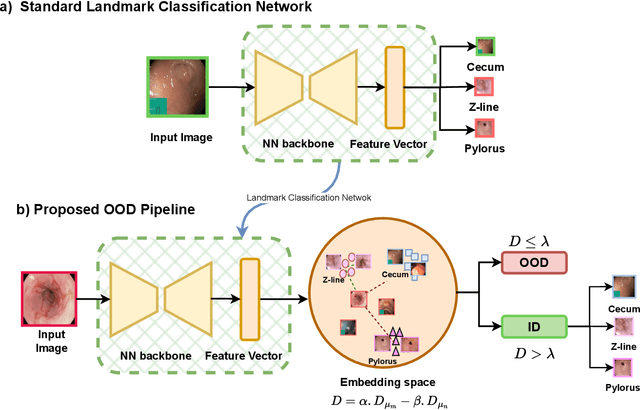
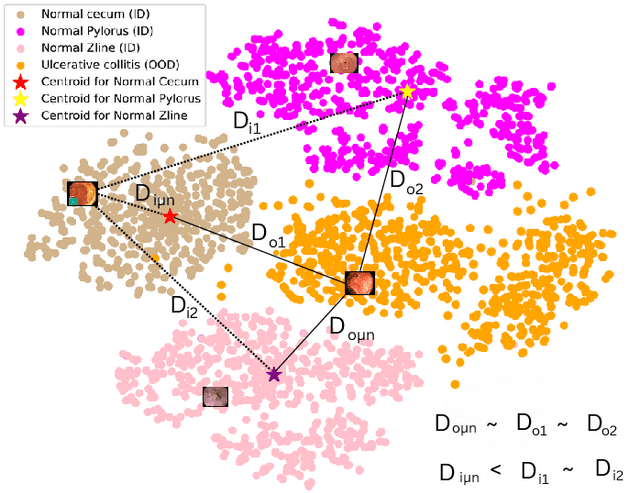
Abstract:The integration of deep learning tools in gastrointestinal vision holds the potential for significant advancements in diagnosis, treatment, and overall patient care. A major challenge, however, is these tools' tendency to make overconfident predictions, even when encountering unseen or newly emerging disease patterns, undermining their reliability. We address this critical issue of reliability by framing it as an out-of-distribution (OOD) detection problem, where previously unseen and emerging diseases are identified as OOD examples. However, gastrointestinal images pose a unique challenge due to the overlapping feature representations between in- Distribution (ID) and OOD examples. Existing approaches often overlook this characteristic, as they are primarily developed for natural image datasets, where feature distinctions are more apparent. Despite the overlap, we hypothesize that the features of an in-distribution example will cluster closer to the centroids of their ground truth class, resulting in a shorter distance to the nearest centroid. In contrast, OOD examples maintain an equal distance from all class centroids. Based on this observation, we propose a novel nearest-centroid distance deficit (NCCD) score in the feature space for gastrointestinal OOD detection. Evaluations across multiple deep learning architectures and two publicly available benchmarks, Kvasir2 and Gastrovision, demonstrate the effectiveness of our approach compared to several state-of-the-art methods. The code and implementation details are publicly available at: https://github.com/bhattarailab/NCDD
Self-supervised Monocular Depth and Pose Estimation for Endoscopy with Generative Latent Priors
Nov 26, 2024


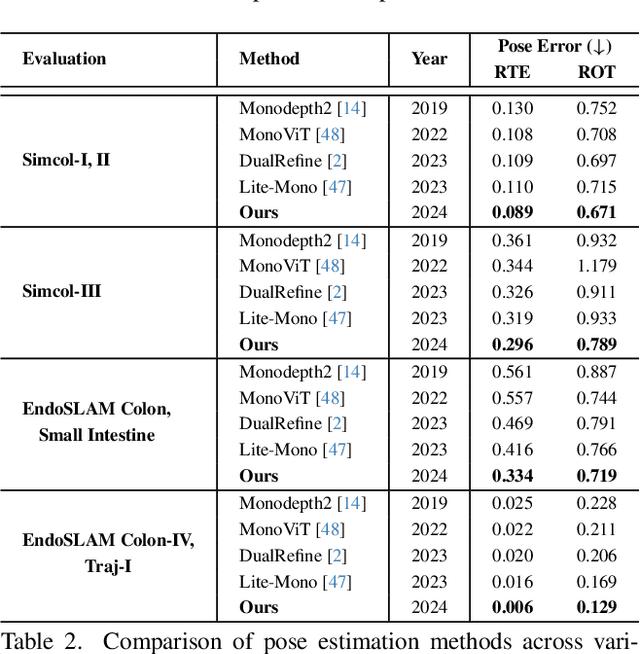
Abstract:Accurate 3D mapping in endoscopy enables quantitative, holistic lesion characterization within the gastrointestinal (GI) tract, requiring reliable depth and pose estimation. However, endoscopy systems are monocular, and existing methods relying on synthetic datasets or complex models often lack generalizability in challenging endoscopic conditions. We propose a robust self-supervised monocular depth and pose estimation framework that incorporates a Generative Latent Bank and a Variational Autoencoder (VAE). The Generative Latent Bank leverages extensive depth scenes from natural images to condition the depth network, enhancing realism and robustness of depth predictions through latent feature priors. For pose estimation, we reformulate it within a VAE framework, treating pose transitions as latent variables to regularize scale, stabilize z-axis prominence, and improve x-y sensitivity. This dual refinement pipeline enables accurate depth and pose predictions, effectively addressing the GI tract's complex textures and lighting. Extensive evaluations on SimCol and EndoSLAM datasets confirm our framework's superior performance over published self-supervised methods in endoscopic depth and pose estimation.
Tackling domain generalization for out-of-distribution endoscopic imaging
Oct 18, 2024Abstract:While recent advances in deep learning (DL) for surgical scene segmentation have yielded promising results on single-center and single-imaging modality data, these methods usually do not generalize well to unseen distributions or modalities. Even though human experts can identify visual appearances, DL methods often fail to do so when data samples do not follow a similar distribution. Current literature addressing domain gaps in modality changes has focused primarily on natural scene data. However, these methods cannot be directly applied to endoscopic data, as visual cues in such data are more limited compared to natural scenes. In this work, we exploit both style and content information in images by performing instance normalization and feature covariance mapping techniques to preserve robust and generalizable feature representations. Additionally, to avoid the risk of removing salient feature representations associated with objects of interest, we introduce a restitution module within the feature-learning ResNet backbone that retains useful task-relevant features. Our proposed method shows a 13.7% improvement over the baseline DeepLabv3+ and nearly an 8% improvement over recent state-of-the-art (SOTA) methods for the target (different modality) set of the EndoUDA polyp dataset. Similarly, our method achieved a 19% improvement over the baseline and 6% over the best-performing SOTA method on the EndoUDA Barrett's esophagus (BE) dataset.
SSTFB: Leveraging self-supervised pretext learning and temporal self-attention with feature branching for real-time video polyp segmentation
Jun 14, 2024Abstract:Polyps are early cancer indicators, so assessing occurrences of polyps and their removal is critical. They are observed through a colonoscopy screening procedure that generates a stream of video frames. Segmenting polyps in their natural video screening procedure has several challenges, such as the co-existence of imaging artefacts, motion blur, and floating debris. Most existing polyp segmentation algorithms are developed on curated still image datasets that do not represent real-world colonoscopy. Their performance often degrades on video data. We propose a video polyp segmentation method that performs self-supervised learning as an auxiliary task and a spatial-temporal self-attention mechanism for improved representation learning. Our end-to-end configuration and joint optimisation of losses enable the network to learn more discriminative contextual features in videos. Our experimental results demonstrate an improvement with respect to several state-of-the-art (SOTA) methods. Our ablation study also confirms that the choice of the proposed joint end-to-end training improves network accuracy by over 3% and nearly 10% on both the Dice similarity coefficient and intersection-over-union compared to the recently proposed method PNS+ and Polyp-PVT, respectively. Results on previously unseen video data indicate that the proposed method generalises.
An objective comparison of methods for augmented reality in laparoscopic liver resection by preoperative-to-intraoperative image fusion
Feb 07, 2024



Abstract:Augmented reality for laparoscopic liver resection is a visualisation mode that allows a surgeon to localise tumours and vessels embedded within the liver by projecting them on top of a laparoscopic image. Preoperative 3D models extracted from CT or MRI data are registered to the intraoperative laparoscopic images during this process. In terms of 3D-2D fusion, most of the algorithms make use of anatomical landmarks to guide registration. These landmarks include the liver's inferior ridge, the falciform ligament, and the occluding contours. They are usually marked by hand in both the laparoscopic image and the 3D model, which is time-consuming and may contain errors if done by a non-experienced user. Therefore, there is a need to automate this process so that augmented reality can be used effectively in the operating room. We present the Preoperative-to-Intraoperative Laparoscopic Fusion Challenge (P2ILF), held during the Medical Imaging and Computer Assisted Interventions (MICCAI 2022) conference, which investigates the possibilities of detecting these landmarks automatically and using them in registration. The challenge was divided into two tasks: 1) A 2D and 3D landmark detection task and 2) a 3D-2D registration task. The teams were provided with training data consisting of 167 laparoscopic images and 9 preoperative 3D models from 9 patients, with the corresponding 2D and 3D landmark annotations. A total of 6 teams from 4 countries participated, whose proposed methods were evaluated on 16 images and two preoperative 3D models from two patients. All the teams proposed deep learning-based methods for the 2D and 3D landmark segmentation tasks and differentiable rendering-based methods for the registration task. Based on the experimental outcomes, we propose three key hypotheses that determine current limitations and future directions for research in this domain.
Multi-task learning with cross-task consistency for improved depth estimation in colonoscopy
Nov 30, 2023



Abstract:Colonoscopy screening is the gold standard procedure for assessing abnormalities in the colon and rectum, such as ulcers and cancerous polyps. Measuring the abnormal mucosal area and its 3D reconstruction can help quantify the surveyed area and objectively evaluate disease burden. However, due to the complex topology of these organs and variable physical conditions, for example, lighting, large homogeneous texture, and image modality estimating distance from the camera aka depth) is highly challenging. Moreover, most colonoscopic video acquisition is monocular, making the depth estimation a non-trivial problem. While methods in computer vision for depth estimation have been proposed and advanced on natural scene datasets, the efficacy of these techniques has not been widely quantified on colonoscopy datasets. As the colonic mucosa has several low-texture regions that are not well pronounced, learning representations from an auxiliary task can improve salient feature extraction, allowing estimation of accurate camera depths. In this work, we propose to develop a novel multi-task learning (MTL) approach with a shared encoder and two decoders, namely a surface normal decoder and a depth estimator decoder. Our depth estimator incorporates attention mechanisms to enhance global context awareness. We leverage the surface normal prediction to improve geometric feature extraction. Also, we apply a cross-task consistency loss among the two geometrically related tasks, surface normal and camera depth. We demonstrate an improvement of 14.17% on relative error and 10.4% improvement on $\delta_{1}$ accuracy over the most accurate baseline state-of-the-art BTS approach. All experiments are conducted on a recently released C3VD dataset; thus, we provide a first benchmark of state-of-the-art methods.
Bayesian uncertainty-weighted loss for improved generalisability on polyp segmentation task
Sep 13, 2023



Abstract:While several previous studies have devised methods for segmentation of polyps, most of these methods are not rigorously assessed on multi-center datasets. Variability due to appearance of polyps from one center to another, difference in endoscopic instrument grades, and acquisition quality result in methods with good performance on in-distribution test data, and poor performance on out-of-distribution or underrepresented samples. Unfair models have serious implications and pose a critical challenge to clinical applications. We adapt an implicit bias mitigation method which leverages Bayesian epistemic uncertainties during training to encourage the model to focus on underrepresented sample regions. We demonstrate the potential of this approach to improve generalisability without sacrificing state-of-the-art performance on a challenging multi-center polyp segmentation dataset (PolypGen) with different centers and image modalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge