Steven A. Hicks
Kvasir-VQA: A Text-Image Pair GI Tract Dataset
Sep 02, 2024



Abstract:We introduce Kvasir-VQA, an extended dataset derived from the HyperKvasir and Kvasir-Instrument datasets, augmented with question-and-answer annotations to facilitate advanced machine learning tasks in Gastrointestinal (GI) diagnostics. This dataset comprises 6,500 annotated images spanning various GI tract conditions and surgical instruments, and it supports multiple question types including yes/no, choice, location, and numerical count. The dataset is intended for applications such as image captioning, Visual Question Answering (VQA), text-based generation of synthetic medical images, object detection, and classification. Our experiments demonstrate the dataset's effectiveness in training models for three selected tasks, showcasing significant applications in medical image analysis and diagnostics. We also present evaluation metrics for each task, highlighting the usability and versatility of our dataset. The dataset and supporting artifacts are available at https://datasets.simula.no/kvasir-vqa.
An objective validation of polyp and instrument segmentation methods in colonoscopy through Medico 2020 polyp segmentation and MedAI 2021 transparency challenges
Jul 30, 2023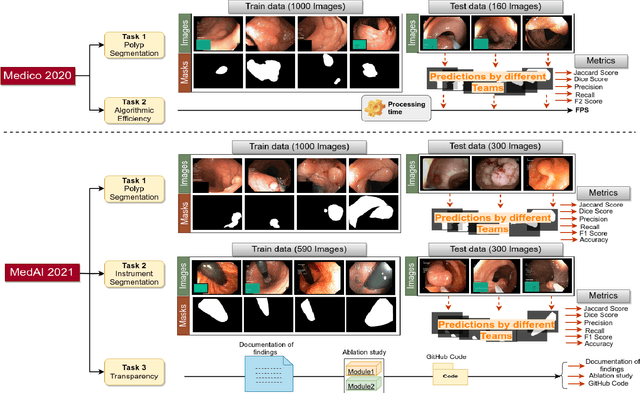
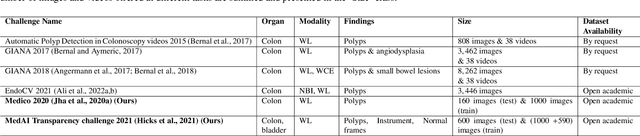
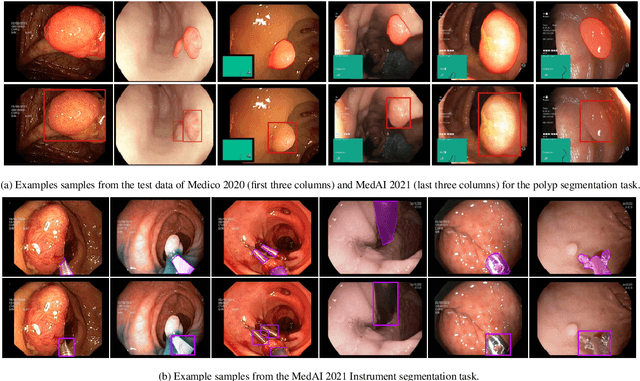
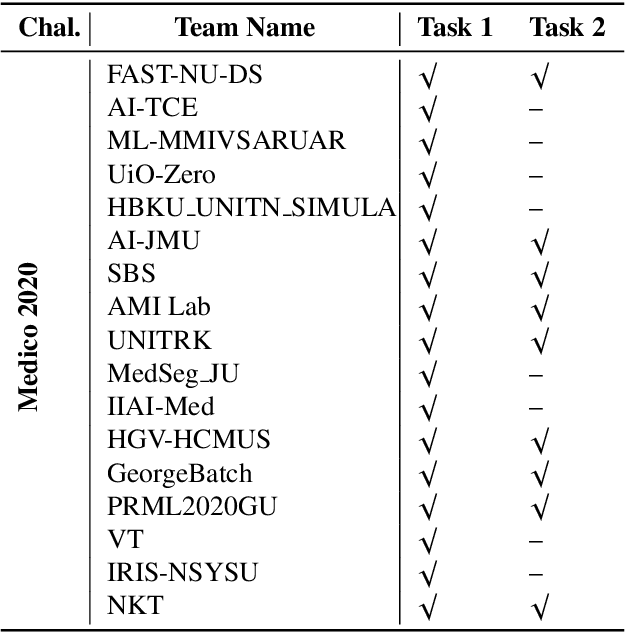
Abstract:Automatic analysis of colonoscopy images has been an active field of research motivated by the importance of early detection of precancerous polyps. However, detecting polyps during the live examination can be challenging due to various factors such as variation of skills and experience among the endoscopists, lack of attentiveness, and fatigue leading to a high polyp miss-rate. Deep learning has emerged as a promising solution to this challenge as it can assist endoscopists in detecting and classifying overlooked polyps and abnormalities in real time. In addition to the algorithm's accuracy, transparency and interpretability are crucial to explaining the whys and hows of the algorithm's prediction. Further, most algorithms are developed in private data, closed source, or proprietary software, and methods lack reproducibility. Therefore, to promote the development of efficient and transparent methods, we have organized the "Medico automatic polyp segmentation (Medico 2020)" and "MedAI: Transparency in Medical Image Segmentation (MedAI 2021)" competitions. We present a comprehensive summary and analyze each contribution, highlight the strength of the best-performing methods, and discuss the possibility of clinical translations of such methods into the clinic. For the transparency task, a multi-disciplinary team, including expert gastroenterologists, accessed each submission and evaluated the team based on open-source practices, failure case analysis, ablation studies, usability and understandability of evaluations to gain a deeper understanding of the models' credibility for clinical deployment. Through the comprehensive analysis of the challenge, we not only highlight the advancements in polyp and surgical instrument segmentation but also encourage qualitative evaluation for building more transparent and understandable AI-based colonoscopy systems.
Blockwise Principal Component Analysis for monotone missing data imputation and dimensionality reduction
May 10, 2023

Abstract:Monotone missing data is a common problem in data analysis. However, imputation combined with dimensionality reduction can be computationally expensive, especially with the increasing size of datasets. To address this issue, we propose a Blockwise principal component analysis Imputation (BPI) framework for dimensionality reduction and imputation of monotone missing data. The framework conducts Principal Component Analysis (PCA) on the observed part of each monotone block of the data and then imputes on merging the obtained principal components using a chosen imputation technique. BPI can work with various imputation techniques and can significantly reduce imputation time compared to conducting dimensionality reduction after imputation. This makes it a practical and efficient approach for large datasets with monotone missing data. Our experiments validate the improvement in speed. In addition, our experiments also show that while applying MICE imputation directly on missing data may not yield convergence, applying BPI with MICE for the data may lead to convergence.
VISEM-Tracking: Human Spermatozoa Tracking Dataset
Dec 23, 2022Abstract:A manual assessment of sperm motility requires microscopy observation, which is challenging due to the fast-moving spermatozoa in the field of view. To obtain correct results, manual evaluation requires extensive training. Therefore, computer-assisted sperm analysis (CASA) has become increasingly used in clinics. Despite this, more data is needed to train supervised machine learning approaches in order to improve accuracy and reliability in the assessment of sperm motility and kinematics. In this regard, we provide a dataset called VISEM-Tracking with 20 video recordings of 30 seconds of wet sperm preparations with manually annotated bounding-box coordinates and a set of sperm characteristics analyzed by experts in the domain. In addition to the annotated data, we provide unlabeled video clips for easy-to-use access and analysis of the data via methods such as self- or unsupervised learning. As part of this paper, we present baseline sperm detection performances using the YOLOv5 deep learning model trained on the VISEM-Tracking dataset. As a result, we show that the dataset can be used to train complex deep learning models to analyze spermatozoa. The dataset is publicly available at https://zenodo.org/record/7293726.
MLC at HECKTOR 2022: The Effect and Importance of Training Data when Analyzing Cases of Head and Neck Tumors using Machine Learning
Nov 30, 2022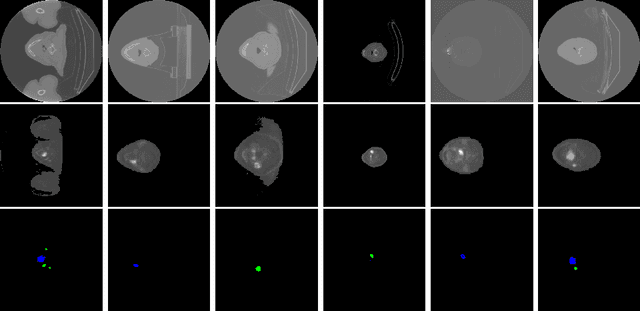
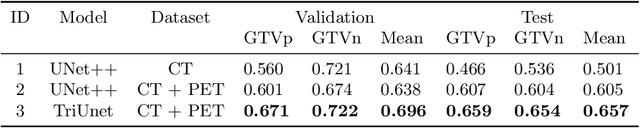
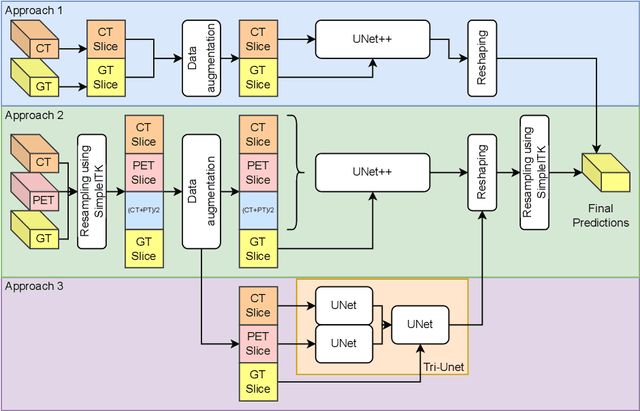
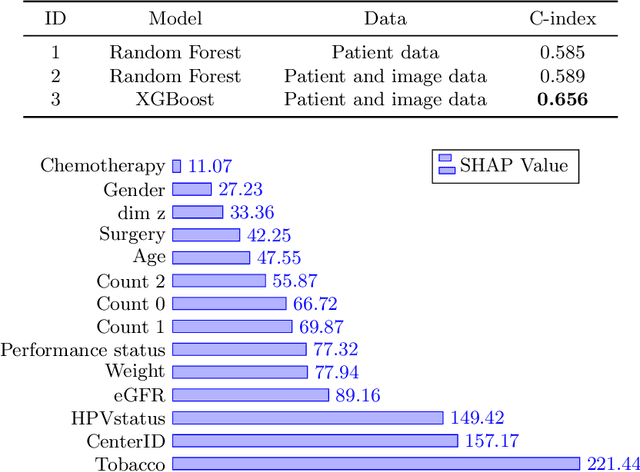
Abstract:Head and neck cancers are the fifth most common cancer worldwide, and recently, analysis of Positron Emission Tomography (PET) and Computed Tomography (CT) images has been proposed to identify patients with a prognosis. Even though the results look promising, more research is needed to further validate and improve the results. This paper presents the work done by team MLC for the 2022 version of the HECKTOR grand challenge held at MICCAI 2022. For Task 1, the automatic segmentation task, our approach was, in contrast to earlier solutions using 3D segmentation, to keep it as simple as possible using a 2D model, analyzing every slice as a standalone image. In addition, we were interested in understanding how different modalities influence the results. We proposed two approaches; one using only the CT scans to make predictions and another using a combination of the CT and PET scans. For Task 2, the prediction of recurrence-free survival, we first proposed two approaches, one where we only use patient data and one where we combined the patient data with segmentations from the image model. For the prediction of the first two approaches, we used Random Forest. In our third approach, we combined patient data and image data using XGBoost. Low kidney function might worsen cancer prognosis. In this approach, we therefore estimated the kidney function of the patients and included it as a feature. Overall, we conclude that our simple methods were not able to compete with the highest-ranking submissions, but we still obtained reasonably good scores. We also got interesting insights into how the combination of different modalities can influence the segmentation and predictions.
MMSys'22 Grand Challenge on AI-based Video Production for Soccer
Feb 02, 2022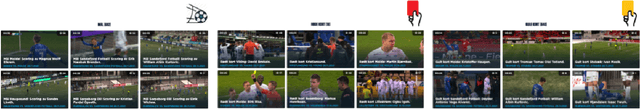
Abstract:Soccer has a considerable market share of the global sports industry, and the interest in viewing videos from soccer games continues to grow. In this respect, it is important to provide game summaries and highlights of the main game events. However, annotating and producing events and summaries often require expensive equipment and a lot of tedious, cumbersome, manual labor. Therefore, automating the video production pipeline providing fast game highlights at a much lower cost is seen as the "holy grail". In this context, recent developments in Artificial Intelligence (AI) technology have shown great potential. Still, state-of-the-art approaches are far from being adequate for practical scenarios that have demanding real-time requirements, as well as strict performance criteria (where at least the detection of official events such as goals and cards must be 100% accurate). In addition, event detection should be thoroughly enhanced by annotation and classification, proper clipping, generating short descriptions, selecting appropriate thumbnails for highlight clips, and finally, combining the event highlights into an overall game summary, similar to what is commonly aired during sports news. Even though the event tagging operation has by far received the most attention, an end-to-end video production pipeline also includes various other operations which serve the overall purpose of automated soccer analysis. This challenge aims to assist the automation of such a production pipeline using AI. In particular, we focus on the enhancement operations that take place after an event has been detected, namely event clipping (Task 1), thumbnail selection (Task 2), and game summarization (Task 3). Challenge website: https://mmsys2022.ie/authors/grand-challenge.
DivergentNets: Medical Image Segmentation by Network Ensemble
Jul 01, 2021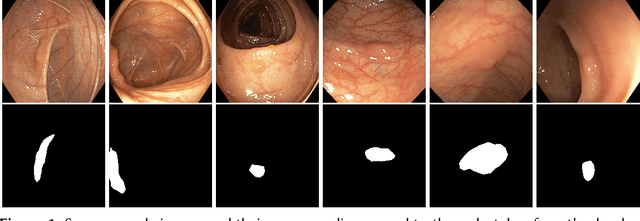
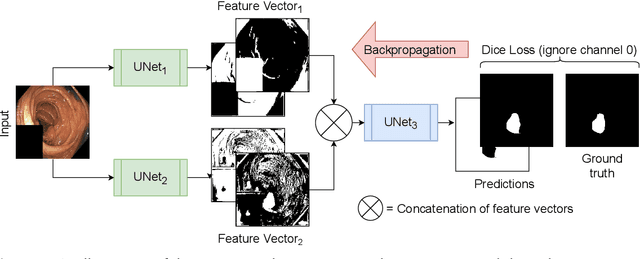
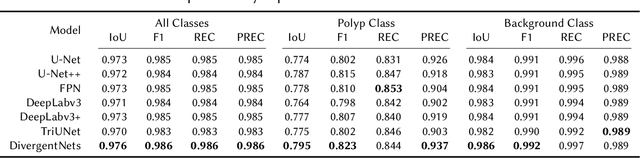
Abstract:Detection of colon polyps has become a trending topic in the intersecting fields of machine learning and gastrointestinal endoscopy. The focus has mainly been on per-frame classification. More recently, polyp segmentation has gained attention in the medical community. Segmentation has the advantage of being more accurate than per-frame classification or object detection as it can show the affected area in greater detail. For our contribution to the EndoCV 2021 segmentation challenge, we propose two separate approaches. First, a segmentation model named TriUNet composed of three separate UNet models. Second, we combine TriUNet with an ensemble of well-known segmentation models, namely UNet++, FPN, DeepLabv3, and DeepLabv3+, into a model called DivergentNets to produce more generalizable medical image segmentation masks. In addition, we propose a modified Dice loss that calculates loss only for a single class when performing multiclass segmentation, forcing the model to focus on what is most important. Overall, the proposed methods achieved the best average scores for each respective round in the challenge, with TriUNet being the winning model in Round I and DivergentNets being the winning model in Round II of the segmentation generalization challenge at EndoCV 2021. The implementation of our approach is made publicly available on GitHub.
* the winning model of the segmentation generalization challenge at EndoCV 2021
SinGAN-Seg: Synthetic Training Data Generation for Medical Image Segmentation
Jun 29, 2021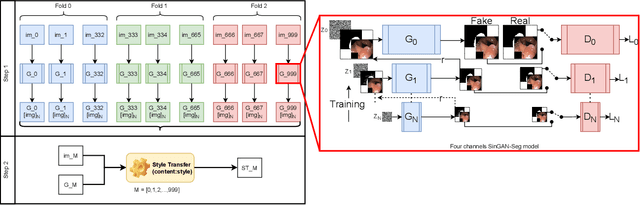
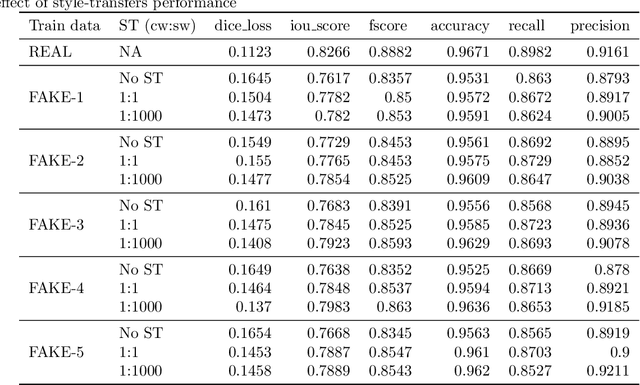
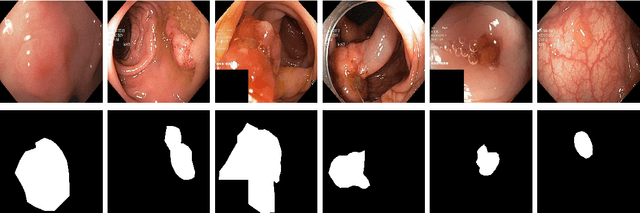
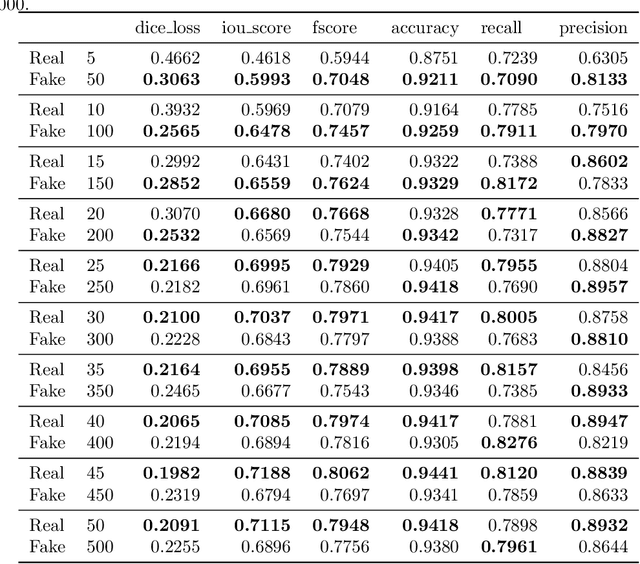
Abstract:Processing medical data to find abnormalities is a time-consuming and costly task, requiring tremendous efforts from medical experts. Therefore, Ai has become a popular tool for the automatic processing of medical data, acting as a supportive tool for doctors. AI tools highly depend on data for training the models. However, there are several constraints to access to large amounts of medical data to train machine learning algorithms in the medical domain, e.g., due to privacy concerns and the costly, time-consuming medical data annotation process. To address this, in this paper we present a novel synthetic data generation pipeline called SinGAN-Seg to produce synthetic medical data with the corresponding annotated ground truth masks. We show that these synthetic data generation pipelines can be used as an alternative to bypass privacy concerns and as an alternative way to produce artificial segmentation datasets with corresponding ground truth masks to avoid the tedious medical data annotation process. As a proof of concept, we used an open polyp segmentation dataset. By training UNet++ using both the real polyp segmentation dataset and the corresponding synthetic dataset generated from the SinGAN-Seg pipeline, we show that the synthetic data can achieve a very close performance to the real data when the real segmentation datasets are large enough. In addition, we show that synthetic data generated from the SinGAN-Seg pipeline improving the performance of segmentation algorithms when the training dataset is very small. Since our SinGAN-Seg pipeline is applicable for any medical dataset, this pipeline can be used with any other segmentation datasets.
Medico Multimedia Task at MediaEval 2020: Automatic Polyp Segmentation
Dec 30, 2020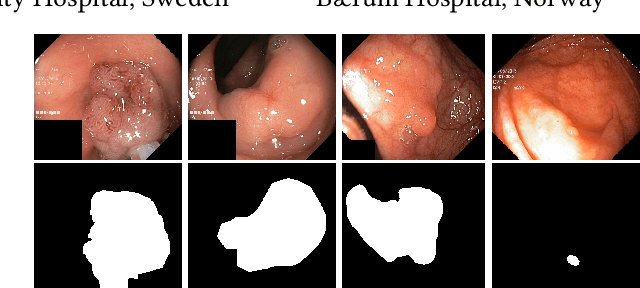
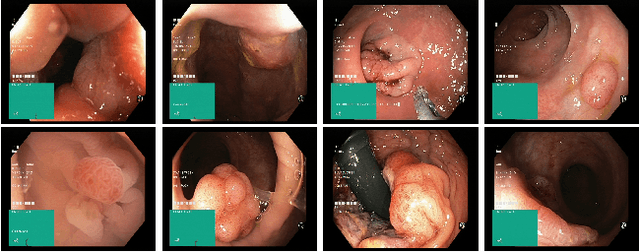
Abstract:Colorectal cancer is the third most common cause of cancer worldwide. According to Global cancer statistics 2018, the incidence of colorectal cancer is increasing in both developing and developed countries. Early detection of colon anomalies such as polyps is important for cancer prevention, and automatic polyp segmentation can play a crucial role for this. Regardless of the recent advancement in early detection and treatment options, the estimated polyp miss rate is still around 20\%. Support via an automated computer-aided diagnosis system could be one of the potential solutions for the overlooked polyps. Such detection systems can help low-cost design solutions and save doctors time, which they could for example use to perform more patient examinations. In this paper, we introduce the 2020 Medico challenge, provide some information on related work and the dataset, describe the task and evaluation metrics, and discuss the necessity of organizing the Medico challenge.
Machine Learning-Based Analysis of Sperm Videos and Participant Data for Male Fertility Prediction
Oct 29, 2019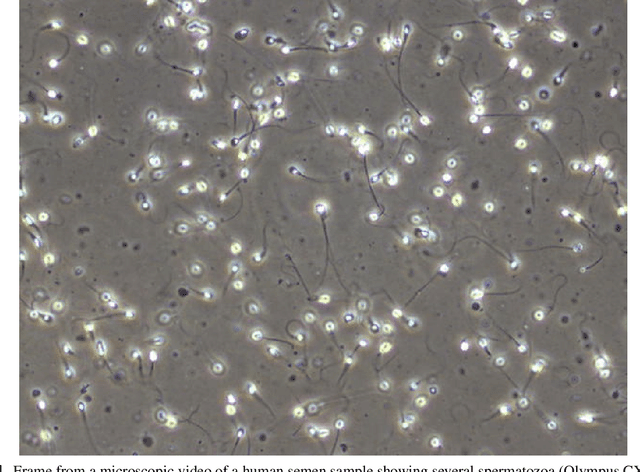
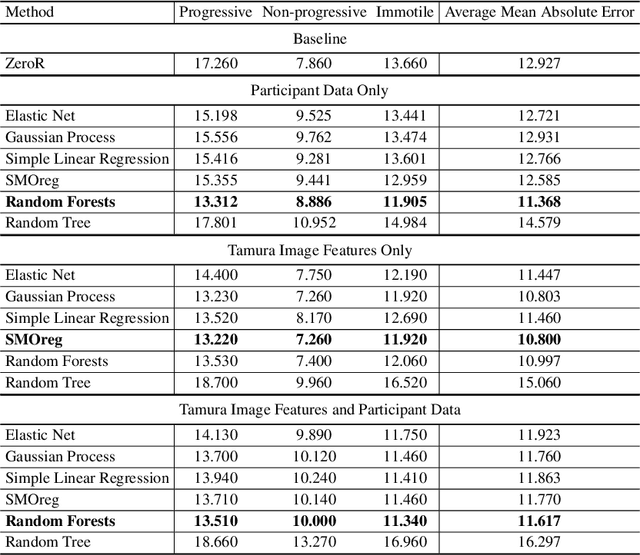
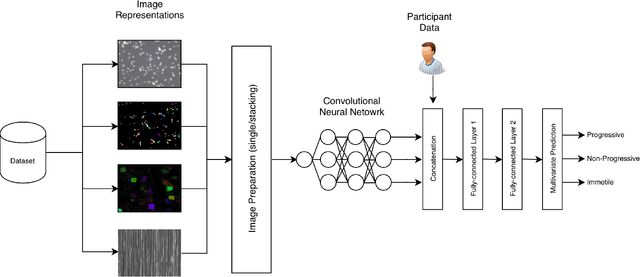
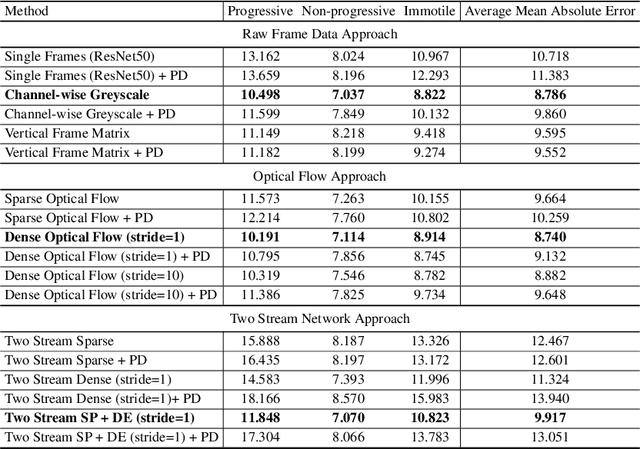
Abstract:Methods for automatic analysis of clinical data are usually targeted towards a specific modality and do not make use of all relevant data available. In the field of male human reproduction, clinical and biological data are not used to its fullest potential. Manual evaluation of a semen sample using a microscope is time-consuming and requires extensive training. Furthermore, the validity of manual semen analysis has been questioned due to limited reproducibility, and often high inter-personnel variation. The existing computer-aided sperm analyzer systems are not recommended for routine clinical use due to methodological challenges caused by the consistency of the semen sample. Thus, there is a need for an improved methodology. We use modern and classical machine learning techniques together with a dataset consisting of 85 videos of human semen samples and related participant data to automatically predict sperm motility. Used techniques include simple linear regression and more sophisticated methods using convolutional neural networks. Our results indicate that sperm motility prediction based on deep learning using sperm motility videos is rapid to perform and consistent. The algorithms performed worse when participant data was added. In conclusion, machine learning-based automatic analysis may become a valuable tool in male infertility investigation and research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge