Vajira Thambawita
Calliope: A TTS-based Narrated E-book Creator Ensuring Exact Synchronization, Privacy, and Layout Fidelity
Feb 11, 2026Abstract:A narrated e-book combines synchronized audio with digital text, highlighting the currently spoken word or sentence during playback. This format supports early literacy and assists individuals with reading challenges, while also allowing general readers to seamlessly switch between reading and listening. With the emergence of natural-sounding neural Text-to-Speech (TTS) technology, several commercial services have been developed to leverage these technology for converting standard text e-books into high-quality narrated e-books. However, no open-source solutions currently exist to perform this task. In this paper, we present Calliope, an open-source framework designed to fill this gap. Our method leverages state-of-the-art open-source TTS to convert a text e-book into a narrated e-book in the EPUB 3 Media Overlay format. The method offers several innovative steps: audio timestamps are captured directly during TTS, ensuring exact synchronization between narration and text highlighting; the publisher's original typography, styling, and embedded media are strictly preserved; and the entire pipeline operates offline. This offline capability eliminates recurring API costs, mitigates privacy concerns, and avoids copyright compliance issues associated with cloud-based services. The framework currently supports the state-of-the-art open-source TTS systems XTTS-v2 and Chatterbox. A potential alternative approach involves first generating narration via TTS and subsequently synchronizing it with the text using forced alignment. However, while our method ensures exact synchronization, our experiments show that forced alignment introduces drift between the audio and text highlighting significant enough to degrade the reading experience. Source code and usage instructions are available at https://github.com/hugohammer/TTS-Narrated-Ebook-Creator.git.
ECG-IMN: Interpretable Mesomorphic Neural Networks for 12-Lead Electrocardiogram Interpretation
Feb 10, 2026Abstract:Deep learning has achieved expert-level performance in automated electrocardiogram (ECG) diagnosis, yet the "black-box" nature of these models hinders their clinical deployment. Trust in medical AI requires not just high accuracy but also transparency regarding the specific physiological features driving predictions. Existing explainability methods for ECGs typically rely on post-hoc approximations (e.g., Grad-CAM and SHAP), which can be unstable, computationally expensive, and unfaithful to the model's actual decision-making process. In this work, we propose the ECG-IMN, an Interpretable Mesomorphic Neural Network tailored for high-resolution 12-lead ECG classification. Unlike standard classifiers, the ECG-IMN functions as a hypernetwork: a deep convolutional backbone generates the parameters of a strictly linear model specific to each input sample. This architecture enforces intrinsic interpretability, as the decision logic is mathematically transparent and the generated weights (W) serve as exact, high-resolution feature attribution maps. We introduce a transition decoder that effectively maps latent features to sample-wise weights, enabling precise localization of pathological evidence (e.g., ST-elevation, T-wave inversion) in both time and lead dimensions. We evaluate our approach on the PTB-XL dataset for classification tasks, demonstrating that the ECG-IMN achieves competitive predictive performance (AUROC comparable to black-box baselines) while providing faithful, instance-specific explanations. By explicitly decoupling parameter generation from prediction execution, our framework bridges the gap between deep learning capability and clinical trustworthiness, offering a principled path toward "white-box" cardiac diagnostics.
Anatomy-Preserving Latent Diffusion for Generation of Brain Segmentation Masks with Ischemic Infarct
Feb 10, 2026Abstract:The scarcity of high-quality segmentation masks remains a major bottleneck for medical image analysis, particularly in non-contrast CT (NCCT) neuroimaging, where manual annotation is costly and variable. To address this limitation, we propose an anatomy-preserving generative framework for the unconditional synthesis of multi-class brain segmentation masks, including ischemic infarcts. The proposed approach combines a variational autoencoder trained exclusively on segmentation masks to learn an anatomical latent representation, with a diffusion model operating in this latent space to generate new samples from pure noise. At inference, synthetic masks are obtained by decoding denoised latent vectors through the frozen VAE decoder, with optional coarse control over lesion presence via a binary prompt. Qualitative results show that the generated masks preserve global brain anatomy, discrete tissue semantics, and realistic variability, while avoiding the structural artifacts commonly observed in pixel-space generative models. Overall, the proposed framework offers a simple and scalable solution for anatomy-aware mask generation in data-scarce medical imaging scenarios.
VideoHEDGE: Entropy-Based Hallucination Detection for Video-VLMs via Semantic Clustering and Spatiotemporal Perturbations
Jan 13, 2026Abstract:Hallucinations in video-capable vision-language models (Video-VLMs) remain frequent and high-confidence, while existing uncertainty metrics often fail to align with correctness. We introduce VideoHEDGE, a modular framework for hallucination detection in video question answering that extends entropy-based reliability estimation from images to temporally structured inputs. Given a video-question pair, VideoHEDGE draws a baseline answer and multiple high-temperature generations from both clean clips and photometrically and spatiotemporally perturbed variants, then clusters the resulting textual outputs into semantic hypotheses using either Natural Language Inference (NLI)-based or embedding-based methods. Cluster-level probability masses yield three reliability scores: Semantic Entropy (SE), RadFlag, and Vision-Amplified Semantic Entropy (VASE). We evaluate VideoHEDGE on the SoccerChat benchmark using an LLM-as-a-judge to obtain binary hallucination labels. Across three 7B Video-VLMs (Qwen2-VL, Qwen2.5-VL, and a SoccerChat-finetuned model), VASE consistently achieves the highest ROC-AUC, especially at larger distortion budgets, while SE and RadFlag often operate near chance. We further show that embedding-based clustering matches NLI-based clustering in detection performance at substantially lower computational cost, and that domain fine-tuning reduces hallucination frequency but yields only modest improvements in calibration. The hedge-bench PyPI library enables reproducible and extensible benchmarking, with full code and experimental resources available at https://github.com/Simula/HEDGE#videohedge .
Medical Imaging AI Competitions Lack Fairness
Dec 19, 2025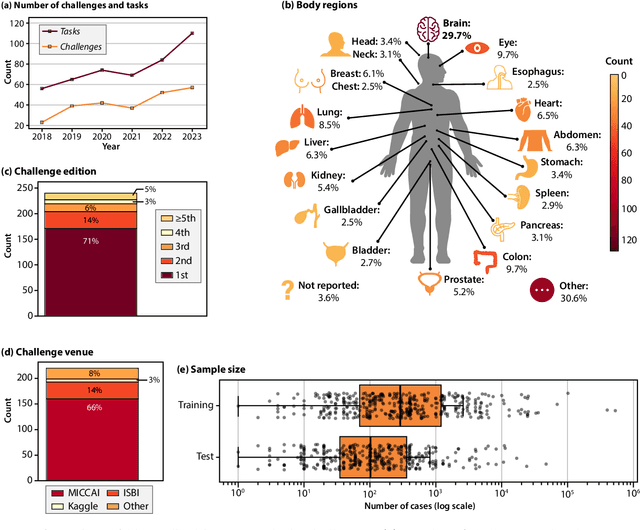
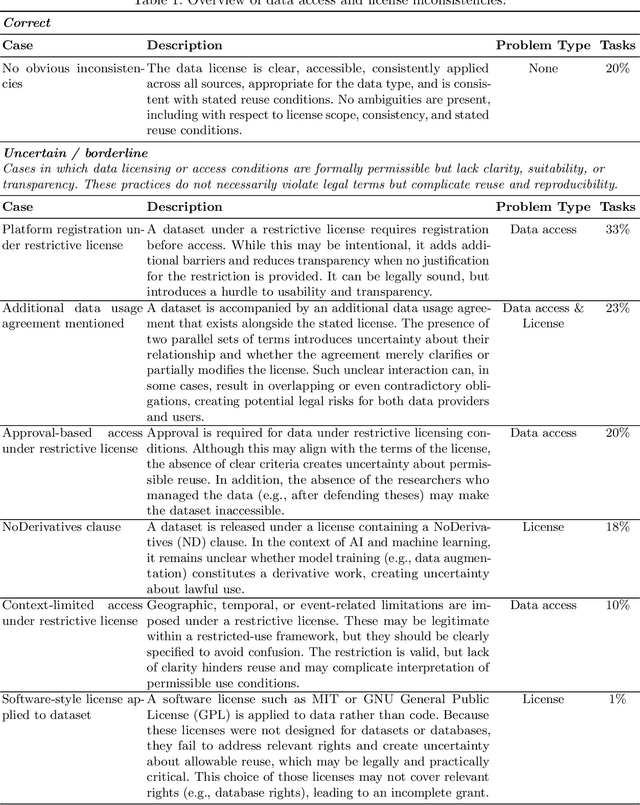
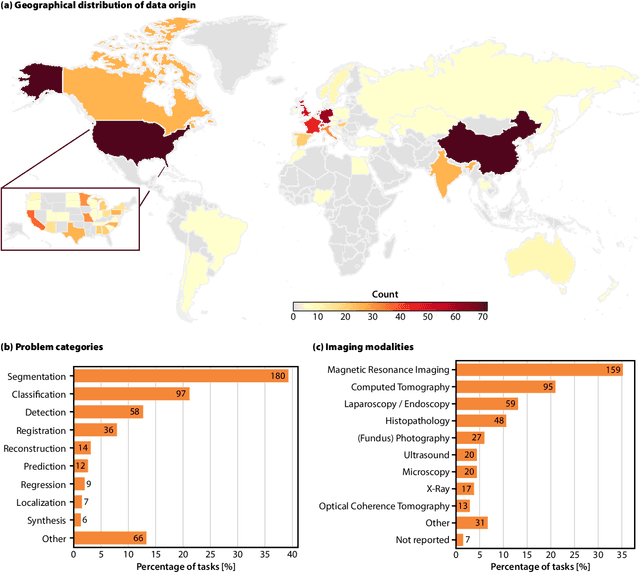
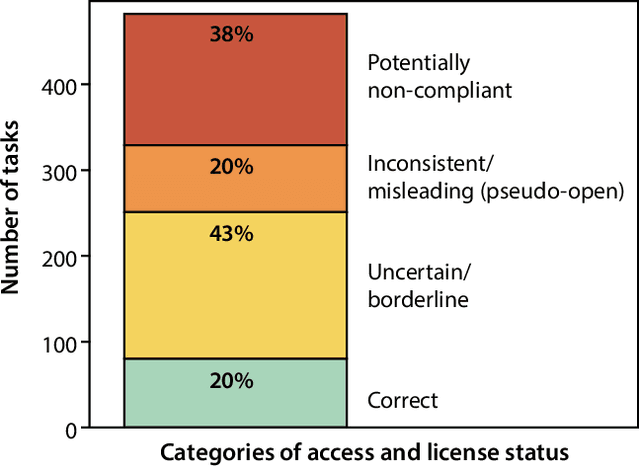
Abstract:Benchmarking competitions are central to the development of artificial intelligence (AI) in medical imaging, defining performance standards and shaping methodological progress. However, it remains unclear whether these benchmarks provide data that are sufficiently representative, accessible, and reusable to support clinically meaningful AI. In this work, we assess fairness along two complementary dimensions: (1) whether challenge datasets are representative of real-world clinical diversity, and (2) whether they are accessible and legally reusable in line with the FAIR principles. To address this question, we conducted a large-scale systematic study of 241 biomedical image analysis challenges comprising 458 tasks across 19 imaging modalities. Our findings show substantial biases in dataset composition, including geographic location, modality-, and problem type-related biases, indicating that current benchmarks do not adequately reflect real-world clinical diversity. Despite their widespread influence, challenge datasets were frequently constrained by restrictive or ambiguous access conditions, inconsistent or non-compliant licensing practices, and incomplete documentation, limiting reproducibility and long-term reuse. Together, these shortcomings expose foundational fairness limitations in our benchmarking ecosystem and highlight a disconnect between leaderboard success and clinical relevance.
From Flat to Feeling: A Feasibility and Impact Study on Dynamic Facial Emotions in AI-Generated Avatars
Jun 16, 2025
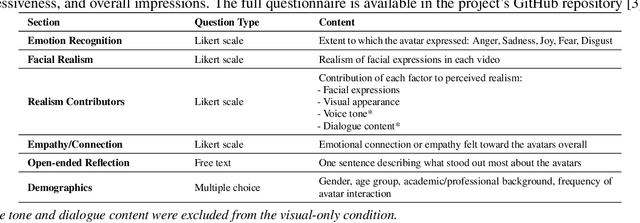
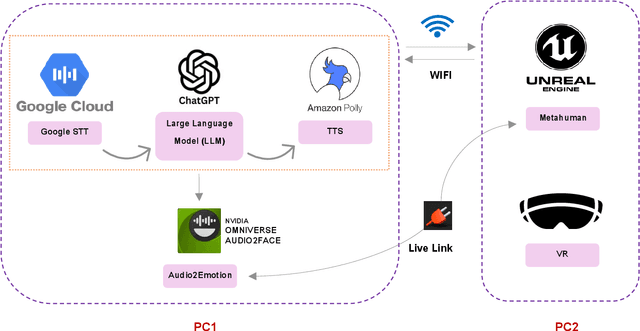

Abstract:Dynamic facial emotion is essential for believable AI-generated avatars; however, most systems remain visually inert, limiting their utility in high-stakes simulations such as virtual training for investigative interviews with abused children. We introduce and evaluate a real-time architecture fusing Unreal Engine 5 MetaHuman rendering with NVIDIA Omniverse Audio2Face to translate vocal prosody into high-fidelity facial expressions on photorealistic child avatars. We implemented a distributed two-PC setup that decouples language processing and speech synthesis from GPU-intensive rendering, designed to support low-latency interaction in desktop and VR environments. A between-subjects study ($N=70$) using audio+visual and visual-only conditions assessed perceptual impacts as participants rated emotional clarity, facial realism, and empathy for two avatars expressing joy, sadness, and anger. Results demonstrate that avatars could express emotions recognizably, with sadness and joy achieving high identification rates. However, anger recognition significantly dropped without audio, highlighting the importance of congruent vocal cues for high-arousal emotions. Interestingly, removing audio boosted perceived facial realism, suggesting that audiovisual desynchrony remains a key design challenge. These findings confirm the technical feasibility of generating emotionally expressive avatars and provide guidance for improving non-verbal communication in sensitive training simulations.
SoccerChat: Integrating Multimodal Data for Enhanced Soccer Game Understanding
May 22, 2025Abstract:The integration of artificial intelligence in sports analytics has transformed soccer video understanding, enabling real-time, automated insights into complex game dynamics. Traditional approaches rely on isolated data streams, limiting their effectiveness in capturing the full context of a match. To address this, we introduce SoccerChat, a multimodal conversational AI framework that integrates visual and textual data for enhanced soccer video comprehension. Leveraging the extensive SoccerNet dataset, enriched with jersey color annotations and automatic speech recognition (ASR) transcripts, SoccerChat is fine-tuned on a structured video instruction dataset to facilitate accurate game understanding, event classification, and referee decision making. We benchmark SoccerChat on action classification and referee decision-making tasks, demonstrating its performance in general soccer event comprehension while maintaining competitive accuracy in referee decision making. Our findings highlight the importance of multimodal integration in advancing soccer analytics, paving the way for more interactive and explainable AI-driven sports analysis. https://github.com/simula/SoccerChat
Embryo 2.0: Merging Synthetic and Real Data for Advanced AI Predictions
Dec 02, 2024Abstract:Accurate embryo morphology assessment is essential in assisted reproductive technology for selecting the most viable embryo. Artificial intelligence has the potential to enhance this process. However, the limited availability of embryo data presents challenges for training deep learning models. To address this, we trained two generative models using two datasets, one we created and made publicly available, and one existing public dataset, to generate synthetic embryo images at various cell stages, including 2-cell, 4-cell, 8-cell, morula, and blastocyst. These were combined with real images to train classification models for embryo cell stage prediction. Our results demonstrate that incorporating synthetic images alongside real data improved classification performance, with the model achieving 97% accuracy compared to 95% when trained solely on real data. Notably, even when trained exclusively on synthetic data and tested on real data, the model achieved a high accuracy of 94%. Furthermore, combining synthetic data from both generative models yielded better classification results than using data from a single generative model. Four embryologists evaluated the fidelity of the synthetic images through a Turing test, during which they annotated inaccuracies and offered feedback. The analysis showed the diffusion model outperformed the generative adversarial network model, deceiving embryologists 66.6% versus 25.3% and achieving lower Frechet inception distance scores.
Comparative Analysis of Audio Feature Extraction for Real-Time Talking Portrait Synthesis
Nov 20, 2024



Abstract:This paper examines the integration of real-time talking-head generation for interviewer training, focusing on overcoming challenges in Audio Feature Extraction (AFE), which often introduces latency and limits responsiveness in real-time applications. To address these issues, we propose and implement a fully integrated system that replaces conventional AFE models with Open AI's Whisper, leveraging its encoder to optimize processing and improve overall system efficiency. Our evaluation of two open-source real-time models across three different datasets shows that Whisper not only accelerates processing but also improves specific aspects of rendering quality, resulting in more realistic and responsive talking-head interactions. These advancements make the system a more effective tool for immersive, interactive training applications, expanding the potential of AI-driven avatars in interviewer training.
Kvasir-VQA: A Text-Image Pair GI Tract Dataset
Sep 02, 2024



Abstract:We introduce Kvasir-VQA, an extended dataset derived from the HyperKvasir and Kvasir-Instrument datasets, augmented with question-and-answer annotations to facilitate advanced machine learning tasks in Gastrointestinal (GI) diagnostics. This dataset comprises 6,500 annotated images spanning various GI tract conditions and surgical instruments, and it supports multiple question types including yes/no, choice, location, and numerical count. The dataset is intended for applications such as image captioning, Visual Question Answering (VQA), text-based generation of synthetic medical images, object detection, and classification. Our experiments demonstrate the dataset's effectiveness in training models for three selected tasks, showcasing significant applications in medical image analysis and diagnostics. We also present evaluation metrics for each task, highlighting the usability and versatility of our dataset. The dataset and supporting artifacts are available at https://datasets.simula.no/kvasir-vqa.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge