Hugo L. Hammer
Calliope: A TTS-based Narrated E-book Creator Ensuring Exact Synchronization, Privacy, and Layout Fidelity
Feb 11, 2026Abstract:A narrated e-book combines synchronized audio with digital text, highlighting the currently spoken word or sentence during playback. This format supports early literacy and assists individuals with reading challenges, while also allowing general readers to seamlessly switch between reading and listening. With the emergence of natural-sounding neural Text-to-Speech (TTS) technology, several commercial services have been developed to leverage these technology for converting standard text e-books into high-quality narrated e-books. However, no open-source solutions currently exist to perform this task. In this paper, we present Calliope, an open-source framework designed to fill this gap. Our method leverages state-of-the-art open-source TTS to convert a text e-book into a narrated e-book in the EPUB 3 Media Overlay format. The method offers several innovative steps: audio timestamps are captured directly during TTS, ensuring exact synchronization between narration and text highlighting; the publisher's original typography, styling, and embedded media are strictly preserved; and the entire pipeline operates offline. This offline capability eliminates recurring API costs, mitigates privacy concerns, and avoids copyright compliance issues associated with cloud-based services. The framework currently supports the state-of-the-art open-source TTS systems XTTS-v2 and Chatterbox. A potential alternative approach involves first generating narration via TTS and subsequently synchronizing it with the text using forced alignment. However, while our method ensures exact synchronization, our experiments show that forced alignment introduces drift between the audio and text highlighting significant enough to degrade the reading experience. Source code and usage instructions are available at https://github.com/hugohammer/TTS-Narrated-Ebook-Creator.git.
ECG-IMN: Interpretable Mesomorphic Neural Networks for 12-Lead Electrocardiogram Interpretation
Feb 10, 2026Abstract:Deep learning has achieved expert-level performance in automated electrocardiogram (ECG) diagnosis, yet the "black-box" nature of these models hinders their clinical deployment. Trust in medical AI requires not just high accuracy but also transparency regarding the specific physiological features driving predictions. Existing explainability methods for ECGs typically rely on post-hoc approximations (e.g., Grad-CAM and SHAP), which can be unstable, computationally expensive, and unfaithful to the model's actual decision-making process. In this work, we propose the ECG-IMN, an Interpretable Mesomorphic Neural Network tailored for high-resolution 12-lead ECG classification. Unlike standard classifiers, the ECG-IMN functions as a hypernetwork: a deep convolutional backbone generates the parameters of a strictly linear model specific to each input sample. This architecture enforces intrinsic interpretability, as the decision logic is mathematically transparent and the generated weights (W) serve as exact, high-resolution feature attribution maps. We introduce a transition decoder that effectively maps latent features to sample-wise weights, enabling precise localization of pathological evidence (e.g., ST-elevation, T-wave inversion) in both time and lead dimensions. We evaluate our approach on the PTB-XL dataset for classification tasks, demonstrating that the ECG-IMN achieves competitive predictive performance (AUROC comparable to black-box baselines) while providing faithful, instance-specific explanations. By explicitly decoupling parameter generation from prediction execution, our framework bridges the gap between deep learning capability and clinical trustworthiness, offering a principled path toward "white-box" cardiac diagnostics.
Explainability of Machine Learning Models under Missing Data
Jun 29, 2024



Abstract:Missing data is a prevalent issue that can significantly impair model performance and interpretability. This paper briefly summarizes the development of the field of missing data with respect to Explainable Artificial Intelligence and experimentally investigates the effects of various imputation methods on the calculation of Shapley values, a popular technique for interpreting complex machine learning models. We compare different imputation strategies and assess their impact on feature importance and interaction as determined by Shapley values. Moreover, we also theoretically analyze the effects of missing values on Shapley values. Importantly, our findings reveal that the choice of imputation method can introduce biases that could lead to changes in the Shapley values, thereby affecting the interpretability of the model. Moreover, and that a lower test prediction mean square error (MSE) may not imply a lower MSE in Shapley values and vice versa. Also, while Xgboost is a method that could handle missing data directly, using Xgboost directly on missing data can seriously affect interpretability compared to imputing the data before training Xgboost. This study provides a comprehensive evaluation of imputation methods in the context of model interpretation, offering practical guidance for selecting appropriate techniques based on dataset characteristics and analysis objectives. The results underscore the importance of considering imputation effects to ensure robust and reliable insights from machine learning models.
Advancing sleep detection by modelling weak label sets: A novel weakly supervised learning approach
Feb 27, 2024



Abstract:Understanding sleep and activity patterns plays a crucial role in physical and mental health. This study introduces a novel approach for sleep detection using weakly supervised learning for scenarios where reliable ground truth labels are unavailable. The proposed method relies on a set of weak labels, derived from the predictions generated by conventional sleep detection algorithms. Introducing a novel approach, we suggest a novel generalised non-linear statistical model in which the number of weak sleep labels is modelled as outcome of a binomial distribution. The probability of sleep in the binomial distribution is linked to the outcomes of neural networks trained to detect sleep based on actigraphy. We show that maximizing the likelihood function of the model, is equivalent to minimizing the soft cross-entropy loss. Additionally, we explored the use of the Brier score as a loss function for weak labels. The efficacy of the suggested modelling framework was demonstrated using the Multi-Ethnic Study of Atherosclerosis dataset. A \gls{lstm} trained on the soft cross-entropy outperformed conventional sleep detection algorithms, other neural network architectures and loss functions in accuracy and model calibration. This research not only advances sleep detection techniques in scenarios where ground truth data is scarce but also contributes to the broader field of weakly supervised learning by introducing innovative approach in modelling sets of weak labels.
VISEM-Tracking: Human Spermatozoa Tracking Dataset
Dec 23, 2022Abstract:A manual assessment of sperm motility requires microscopy observation, which is challenging due to the fast-moving spermatozoa in the field of view. To obtain correct results, manual evaluation requires extensive training. Therefore, computer-assisted sperm analysis (CASA) has become increasingly used in clinics. Despite this, more data is needed to train supervised machine learning approaches in order to improve accuracy and reliability in the assessment of sperm motility and kinematics. In this regard, we provide a dataset called VISEM-Tracking with 20 video recordings of 30 seconds of wet sperm preparations with manually annotated bounding-box coordinates and a set of sperm characteristics analyzed by experts in the domain. In addition to the annotated data, we provide unlabeled video clips for easy-to-use access and analysis of the data via methods such as self- or unsupervised learning. As part of this paper, we present baseline sperm detection performances using the YOLOv5 deep learning model trained on the VISEM-Tracking dataset. As a result, we show that the dataset can be used to train complex deep learning models to analyze spermatozoa. The dataset is publicly available at https://zenodo.org/record/7293726.
Artificial Intelligence in Dry Eye Disease
Sep 02, 2021Abstract:Dry eye disease (DED) has a prevalence of between 5 and 50\%, depending on the diagnostic criteria used and population under study. However, it remains one of the most underdiagnosed and undertreated conditions in ophthalmology. Many tests used in the diagnosis of DED rely on an experienced observer for image interpretation, which may be considered subjective and result in variation in diagnosis. Since artificial intelligence (AI) systems are capable of advanced problem solving, use of such techniques could lead to more objective diagnosis. Although the term `AI' is commonly used, recent success in its applications to medicine is mainly due to advancements in the sub-field of machine learning, which has been used to automatically classify images and predict medical outcomes. Powerful machine learning techniques have been harnessed to understand nuances in patient data and medical images, aiming for consistent diagnosis and stratification of disease severity. This is the first literature review on the use of AI in DED. We provide a brief introduction to AI, report its current use in DED research and its potential for application in the clinic. Our review found that AI has been employed in a wide range of DED clinical tests and research applications, primarily for interpretation of interferometry, slit-lamp and meibography images. While initial results are promising, much work is still needed on model development, clinical testing and standardisation.
SinGAN-Seg: Synthetic Training Data Generation for Medical Image Segmentation
Jun 29, 2021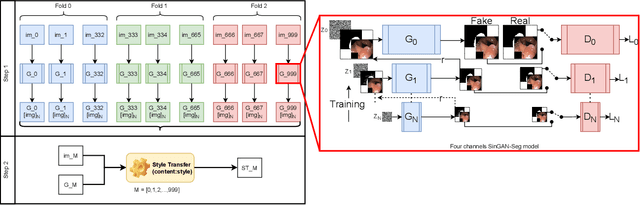
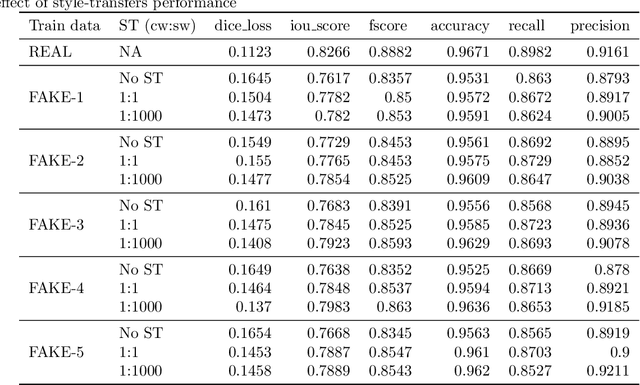
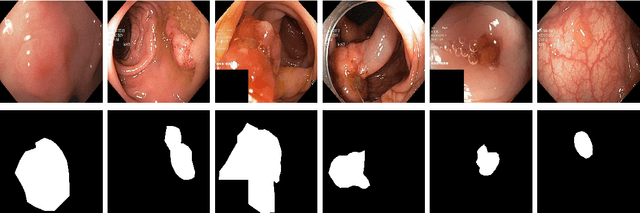
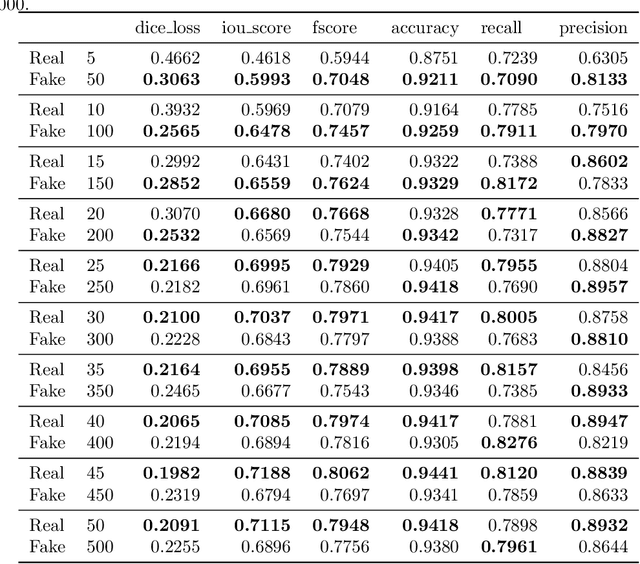
Abstract:Processing medical data to find abnormalities is a time-consuming and costly task, requiring tremendous efforts from medical experts. Therefore, Ai has become a popular tool for the automatic processing of medical data, acting as a supportive tool for doctors. AI tools highly depend on data for training the models. However, there are several constraints to access to large amounts of medical data to train machine learning algorithms in the medical domain, e.g., due to privacy concerns and the costly, time-consuming medical data annotation process. To address this, in this paper we present a novel synthetic data generation pipeline called SinGAN-Seg to produce synthetic medical data with the corresponding annotated ground truth masks. We show that these synthetic data generation pipelines can be used as an alternative to bypass privacy concerns and as an alternative way to produce artificial segmentation datasets with corresponding ground truth masks to avoid the tedious medical data annotation process. As a proof of concept, we used an open polyp segmentation dataset. By training UNet++ using both the real polyp segmentation dataset and the corresponding synthetic dataset generated from the SinGAN-Seg pipeline, we show that the synthetic data can achieve a very close performance to the real data when the real segmentation datasets are large enough. In addition, we show that synthetic data generated from the SinGAN-Seg pipeline improving the performance of segmentation algorithms when the training dataset is very small. Since our SinGAN-Seg pipeline is applicable for any medical dataset, this pipeline can be used with any other segmentation datasets.
Dyadic aggregated autoregressive (DASAR) model for time-frequency representation of biomedical signals
May 13, 2021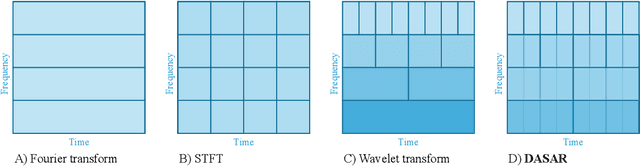
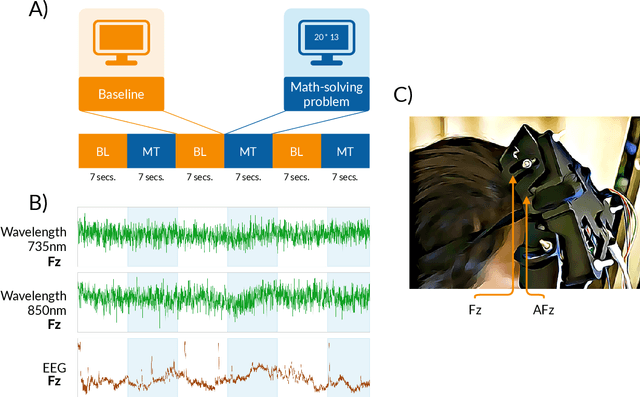
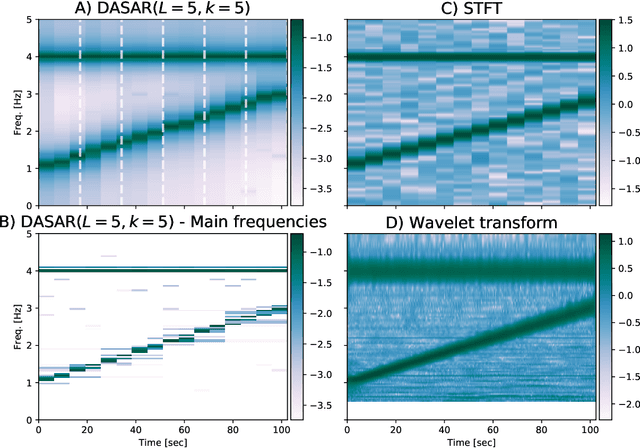
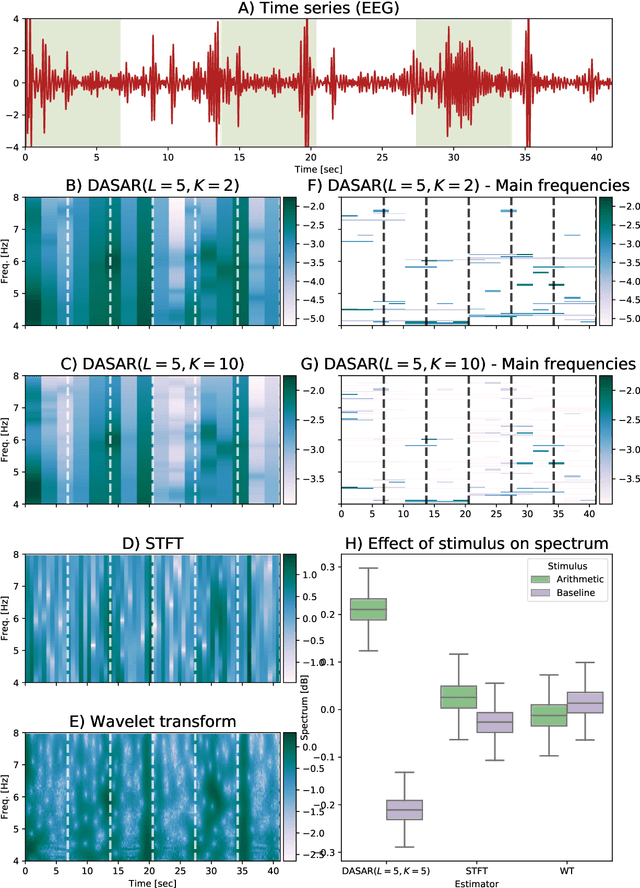
Abstract:This paper introduces a new time-frequency representation method for biomedical signals: the dyadic aggregated autoregressive (DASAR) model. Signals, such as electroencephalograms (EEGs) and functional near-infrared spectroscopy (fNIRS), exhibit physiological information through time-evolving spectrum components at specific frequency intervals: 0-50 Hz (EEG) or 0-150 mHz (fNIRS). Spectrotemporal features in signals are conventionally estimated using short-time Fourier transform (STFT) and wavelet transform (WT). However, both methods may not offer the most robust or compact representation despite their widespread use in biomedical contexts. The presented method, DASAR, improves precise frequency identification and tracking of interpretable frequency components with a parsimonious set of parameters. DASAR achieves these characteristics by assuming that the biomedical time-varying spectrum comprises several independent stochastic oscillators with (piecewise) time-varying frequencies. Local stationarity can be assumed within dyadic subdivisions of the recordings, while the stochastic oscillators can be modeled with an aggregation of second-order autoregressive models (ASAR). DASAR can provide a more accurate representation of the (highly contrasted) EEG and fNIRS frequency ranges by increasing the estimation accuracy in user-defined spectrum region of interest (SROI). A mental arithmetic experiment on a hybrid EEG-fNIRS was conducted to assess the efficiency of the method. Our proposed technique, STFT, and WT were applied on both biomedical signals to discover potential oscillators that improve the discrimination between the task condition and its baseline. The results show that DASAR provided the highest spectrum differentiation and it was the only method that could identify Mayer waves as narrow-band artifacts at 97.4-97.5 mHz.
Efficient Quantile Tracking Using an Oracle
Apr 27, 2020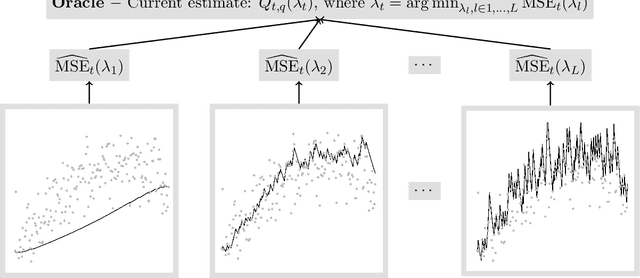
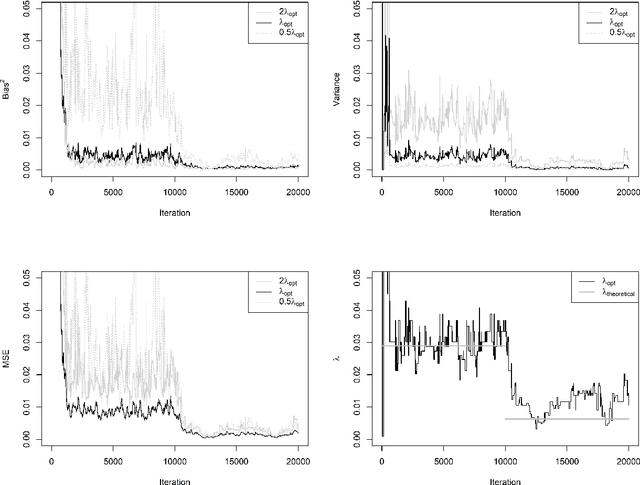
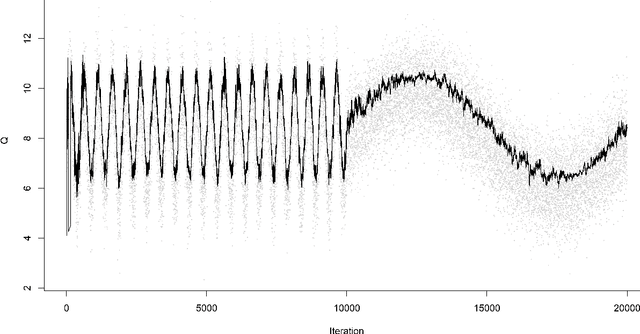
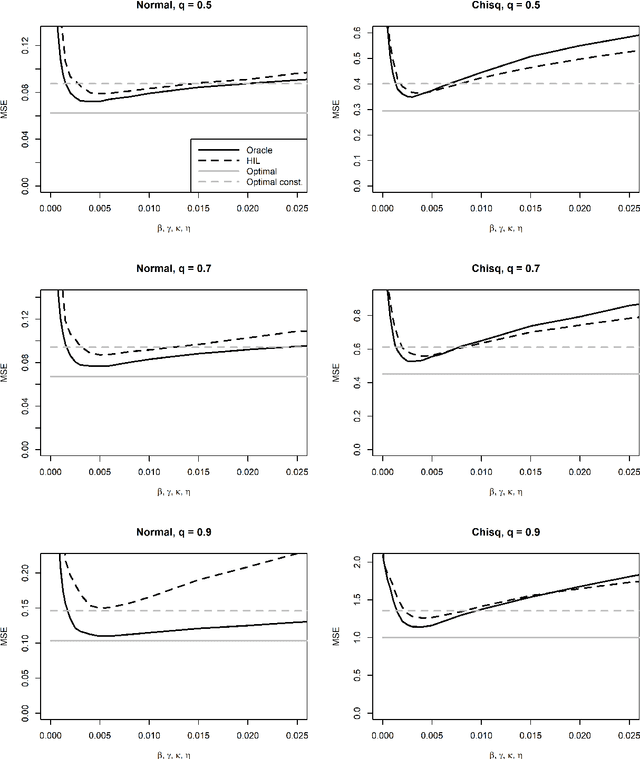
Abstract:For incremental quantile estimators the step size and possibly other tuning parameters must be carefully set. However, little attention has been given on how to set these values in an online manner. In this article we suggest two novel procedures that address this issue. The core part of the procedures is to estimate the current tracking mean squared error (MSE). The MSE is decomposed in tracking variance and bias and novel and efficient procedures to estimate these quantities are presented. It is shown that estimation bias can be tracked by associating it with the portion of observations below the quantile estimates. The first procedure runs an ensemble of $L$ quantile estimators for wide range of values of the tuning parameters and typically around $L = 100$. In each iteration an oracle selects the best estimate by the guidance of the estimated MSEs. The second method only runs an ensemble of $L = 3$ estimators and thus the values of the tuning parameters need from time to time to be adjusted for the running estimators. The procedures have a low memory foot print of $8L$ and a computational complexity of $8L$ per iteration. The experiments show that the procedures are highly efficient and track quantiles with an error close to the theoretical optimum. The Oracle approach performs best, but comes with higher computational cost. The procedures were further applied to a massive real-life data stream of tweets and proofed real world applicability of them.
A hemodynamic decomposition model for detecting cognitive load using functional near-infrared spectroscopy
Jan 22, 2020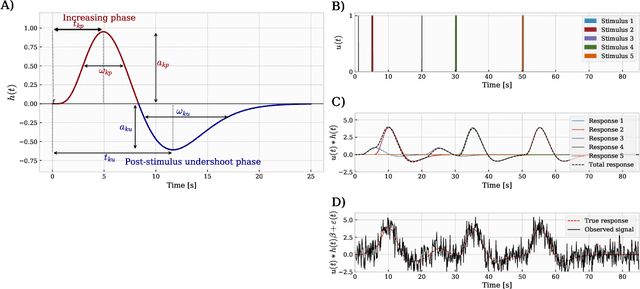
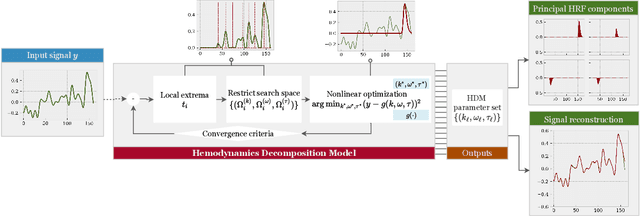
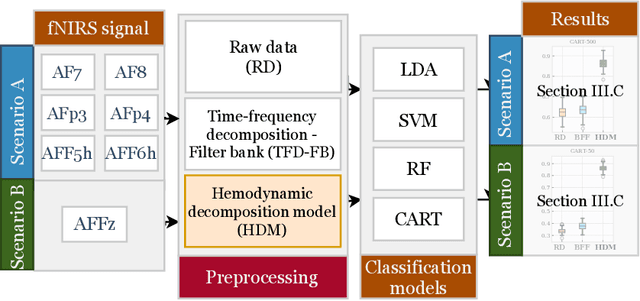
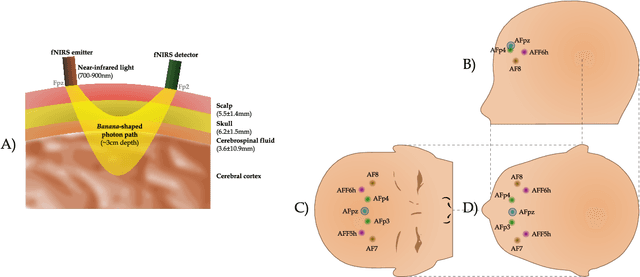
Abstract:In the current paper, we introduce a parametric data-driven model for functional near-infrared spectroscopy that decomposes a signal into a series of independent, rescaled, time-shifted, hemodynamic basis functions. Each decomposed waveform retains relevant biological information about the expected hemodynamic behavior. The model is also presented along with an efficient iterative estimation method to improve the computational speed. Our hemodynamic decomposition model (HDM) extends the canonical model for instances when a) the external stimuli are unknown, or b) when the assumption of a direct relationship between the experimental stimuli and the hemodynamic responses cannot hold. We also argue that the proposed approach can be potentially adopted as a feature transformation method for machine learning purposes. By virtue of applying our devised HDM to a cognitive load classification task on fNIRS signals, we have achieved an accuracy of 86.20%+-2.56% using six channels in the frontal cortex, and 86.34%+-2.81% utilizing only the AFpz channel also located in the frontal area. In comparison, state-of-the-art time-spectral transformations only yield 64.61%+-3.03% and 37.8%+-2.96% under identical experimental settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge