Marco A. Pinto-Orellana
Dyadic aggregated autoregressive (DASAR) model for time-frequency representation of biomedical signals
May 13, 2021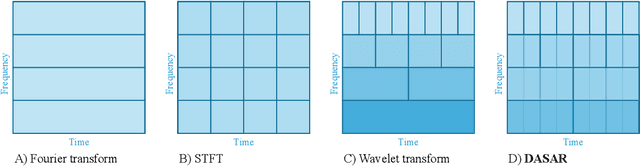
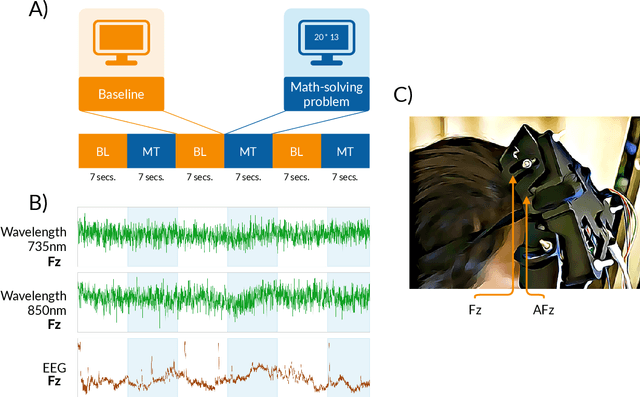
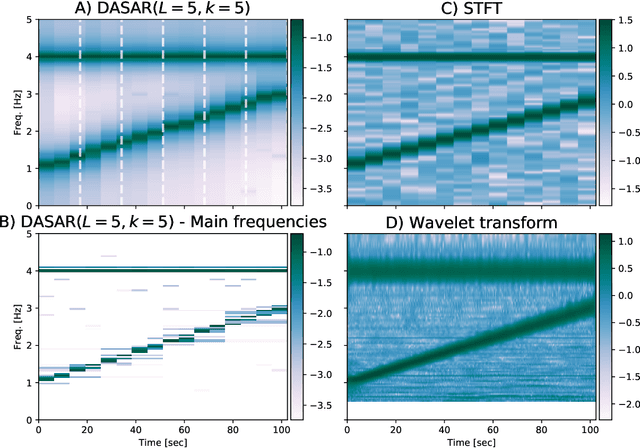
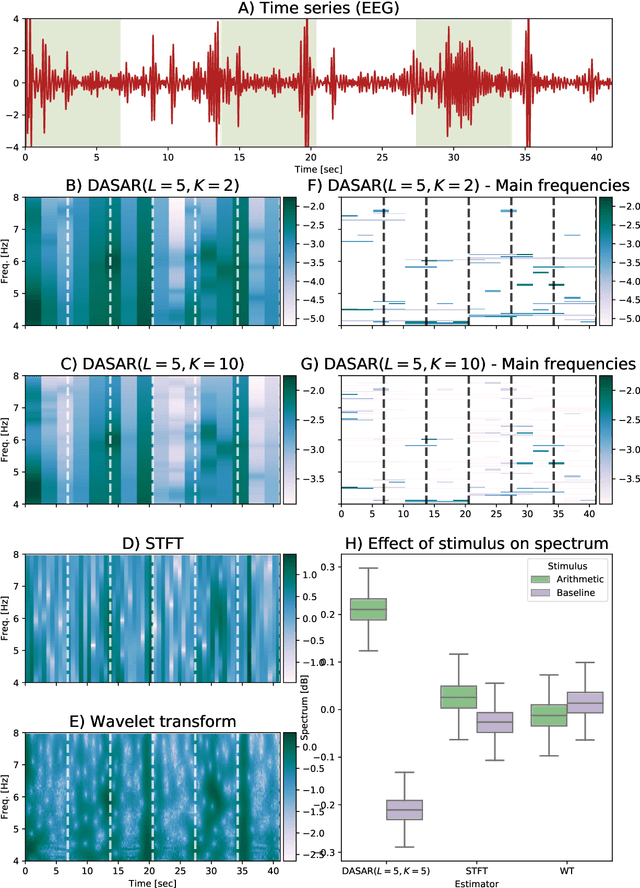
Abstract:This paper introduces a new time-frequency representation method for biomedical signals: the dyadic aggregated autoregressive (DASAR) model. Signals, such as electroencephalograms (EEGs) and functional near-infrared spectroscopy (fNIRS), exhibit physiological information through time-evolving spectrum components at specific frequency intervals: 0-50 Hz (EEG) or 0-150 mHz (fNIRS). Spectrotemporal features in signals are conventionally estimated using short-time Fourier transform (STFT) and wavelet transform (WT). However, both methods may not offer the most robust or compact representation despite their widespread use in biomedical contexts. The presented method, DASAR, improves precise frequency identification and tracking of interpretable frequency components with a parsimonious set of parameters. DASAR achieves these characteristics by assuming that the biomedical time-varying spectrum comprises several independent stochastic oscillators with (piecewise) time-varying frequencies. Local stationarity can be assumed within dyadic subdivisions of the recordings, while the stochastic oscillators can be modeled with an aggregation of second-order autoregressive models (ASAR). DASAR can provide a more accurate representation of the (highly contrasted) EEG and fNIRS frequency ranges by increasing the estimation accuracy in user-defined spectrum region of interest (SROI). A mental arithmetic experiment on a hybrid EEG-fNIRS was conducted to assess the efficiency of the method. Our proposed technique, STFT, and WT were applied on both biomedical signals to discover potential oscillators that improve the discrimination between the task condition and its baseline. The results show that DASAR provided the highest spectrum differentiation and it was the only method that could identify Mayer waves as narrow-band artifacts at 97.4-97.5 mHz.
A hemodynamic decomposition model for detecting cognitive load using functional near-infrared spectroscopy
Jan 22, 2020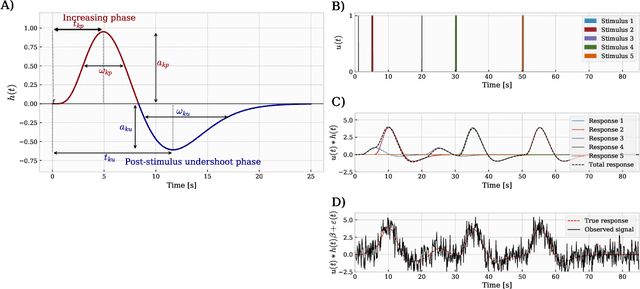
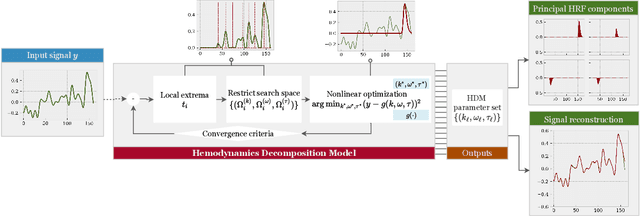
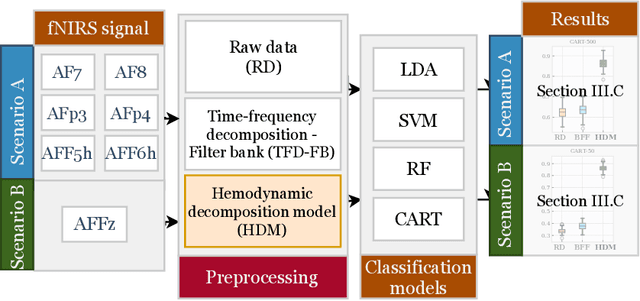
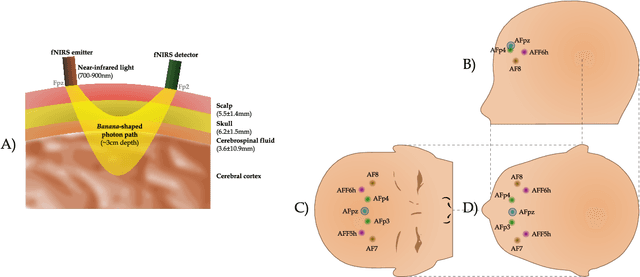
Abstract:In the current paper, we introduce a parametric data-driven model for functional near-infrared spectroscopy that decomposes a signal into a series of independent, rescaled, time-shifted, hemodynamic basis functions. Each decomposed waveform retains relevant biological information about the expected hemodynamic behavior. The model is also presented along with an efficient iterative estimation method to improve the computational speed. Our hemodynamic decomposition model (HDM) extends the canonical model for instances when a) the external stimuli are unknown, or b) when the assumption of a direct relationship between the experimental stimuli and the hemodynamic responses cannot hold. We also argue that the proposed approach can be potentially adopted as a feature transformation method for machine learning purposes. By virtue of applying our devised HDM to a cognitive load classification task on fNIRS signals, we have achieved an accuracy of 86.20%+-2.56% using six channels in the frontal cortex, and 86.34%+-2.81% utilizing only the AFpz channel also located in the frontal area. In comparison, state-of-the-art time-spectral transformations only yield 64.61%+-3.03% and 37.8%+-2.96% under identical experimental settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge