Michael A. Riegler
VideoHEDGE: Entropy-Based Hallucination Detection for Video-VLMs via Semantic Clustering and Spatiotemporal Perturbations
Jan 13, 2026Abstract:Hallucinations in video-capable vision-language models (Video-VLMs) remain frequent and high-confidence, while existing uncertainty metrics often fail to align with correctness. We introduce VideoHEDGE, a modular framework for hallucination detection in video question answering that extends entropy-based reliability estimation from images to temporally structured inputs. Given a video-question pair, VideoHEDGE draws a baseline answer and multiple high-temperature generations from both clean clips and photometrically and spatiotemporally perturbed variants, then clusters the resulting textual outputs into semantic hypotheses using either Natural Language Inference (NLI)-based or embedding-based methods. Cluster-level probability masses yield three reliability scores: Semantic Entropy (SE), RadFlag, and Vision-Amplified Semantic Entropy (VASE). We evaluate VideoHEDGE on the SoccerChat benchmark using an LLM-as-a-judge to obtain binary hallucination labels. Across three 7B Video-VLMs (Qwen2-VL, Qwen2.5-VL, and a SoccerChat-finetuned model), VASE consistently achieves the highest ROC-AUC, especially at larger distortion budgets, while SE and RadFlag often operate near chance. We further show that embedding-based clustering matches NLI-based clustering in detection performance at substantially lower computational cost, and that domain fine-tuning reduces hallucination frequency but yields only modest improvements in calibration. The hedge-bench PyPI library enables reproducible and extensible benchmarking, with full code and experimental resources available at https://github.com/Simula/HEDGE#videohedge .
Medical Imaging AI Competitions Lack Fairness
Dec 19, 2025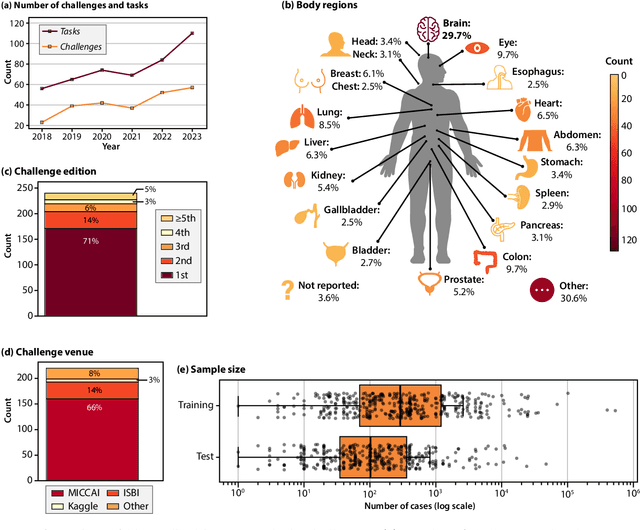
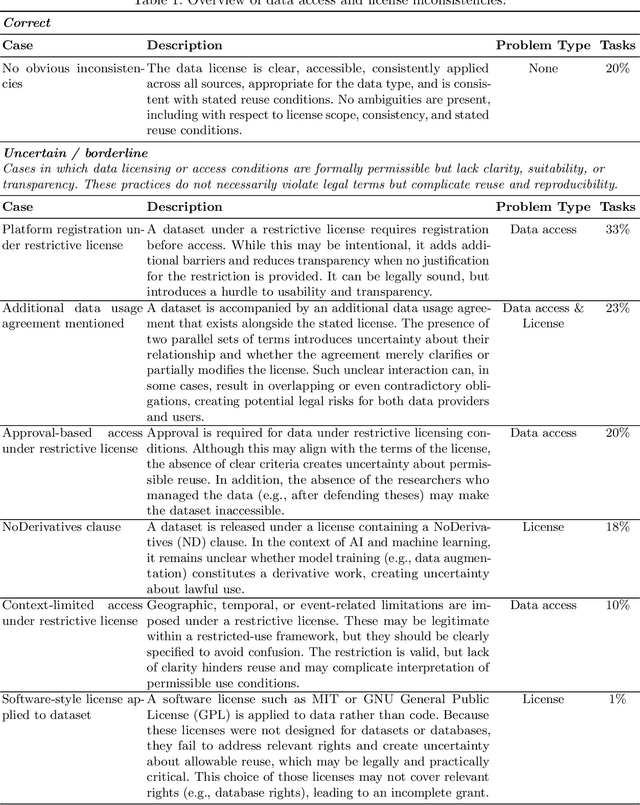
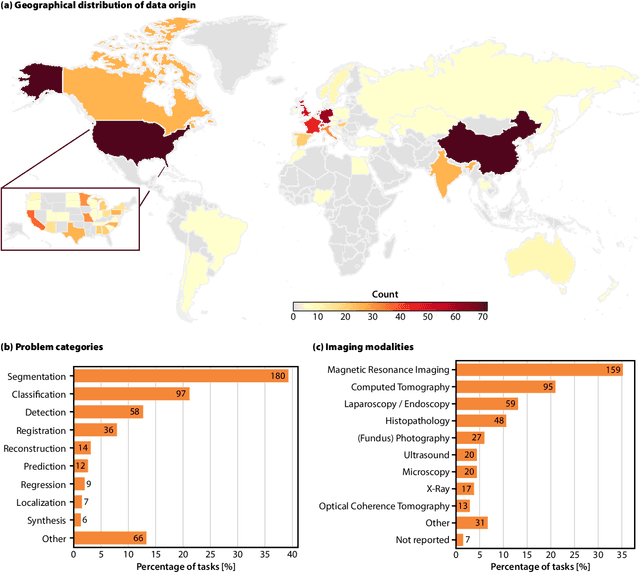
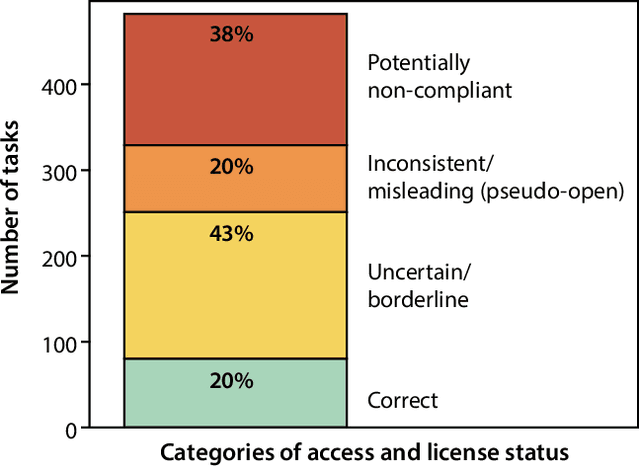
Abstract:Benchmarking competitions are central to the development of artificial intelligence (AI) in medical imaging, defining performance standards and shaping methodological progress. However, it remains unclear whether these benchmarks provide data that are sufficiently representative, accessible, and reusable to support clinically meaningful AI. In this work, we assess fairness along two complementary dimensions: (1) whether challenge datasets are representative of real-world clinical diversity, and (2) whether they are accessible and legally reusable in line with the FAIR principles. To address this question, we conducted a large-scale systematic study of 241 biomedical image analysis challenges comprising 458 tasks across 19 imaging modalities. Our findings show substantial biases in dataset composition, including geographic location, modality-, and problem type-related biases, indicating that current benchmarks do not adequately reflect real-world clinical diversity. Despite their widespread influence, challenge datasets were frequently constrained by restrictive or ambiguous access conditions, inconsistent or non-compliant licensing practices, and incomplete documentation, limiting reproducibility and long-term reuse. Together, these shortcomings expose foundational fairness limitations in our benchmarking ecosystem and highlight a disconnect between leaderboard success and clinical relevance.
HEDGE: Hallucination Estimation via Dense Geometric Entropy for VQA with Vision-Language Models
Nov 16, 2025Abstract:Vision-language models (VLMs) enable open-ended visual question answering but remain prone to hallucinations. We present HEDGE, a unified framework for hallucination detection that combines controlled visual perturbations, semantic clustering, and robust uncertainty metrics. HEDGE integrates sampling, distortion synthesis, clustering (entailment- and embedding-based), and metric computation into a reproducible pipeline applicable across multimodal architectures. Evaluations on VQA-RAD and KvasirVQA-x1 with three representative VLMs (LLaVA-Med, Med-Gemma, Qwen2.5-VL) reveal clear architecture- and prompt-dependent trends. Hallucination detectability is highest for unified-fusion models with dense visual tokenization (Qwen2.5-VL) and lowest for architectures with restricted tokenization (Med-Gemma). Embedding-based clustering often yields stronger separation when applied directly to the generated answers, whereas NLI-based clustering remains advantageous for LLaVA-Med and for longer, sentence-level responses. Across configurations, the VASE metric consistently provides the most robust hallucination signal, especially when paired with embedding clustering and a moderate sampling budget (n ~ 10-15). Prompt design also matters: concise, label-style outputs offer clearer semantic structure than syntactically constrained one-sentence responses. By framing hallucination detection as a geometric robustness problem shaped jointly by sampling scale, prompt structure, model architecture, and clustering strategy, HEDGE provides a principled, compute-aware foundation for evaluating multimodal reliability. The hedge-bench PyPI library enables reproducible and extensible benchmarking, with full code and experimental resources available at https://github.com/Simula/HEDGE .
Kvasir-VQA-x1: A Multimodal Dataset for Medical Reasoning and Robust MedVQA in Gastrointestinal Endoscopy
Jun 11, 2025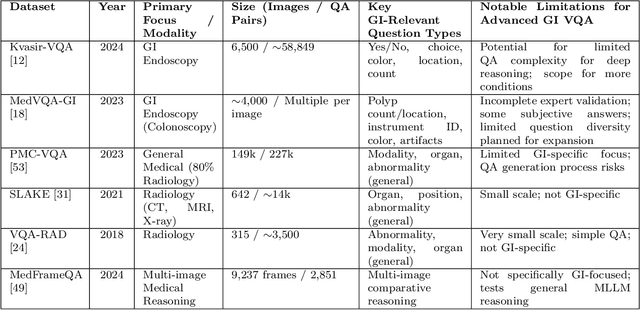
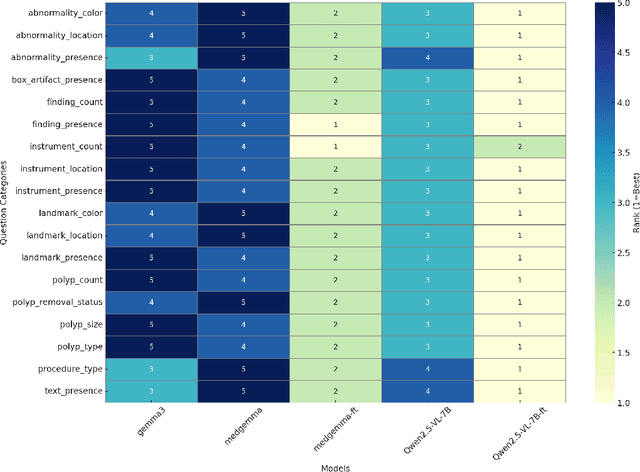
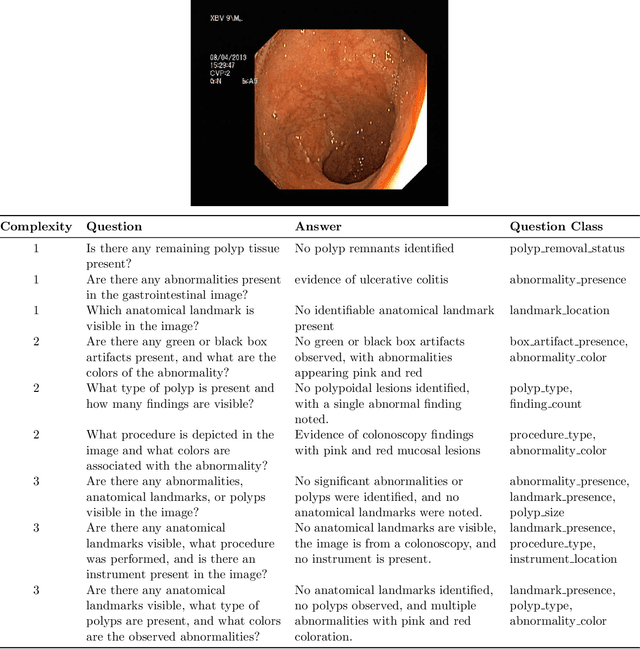
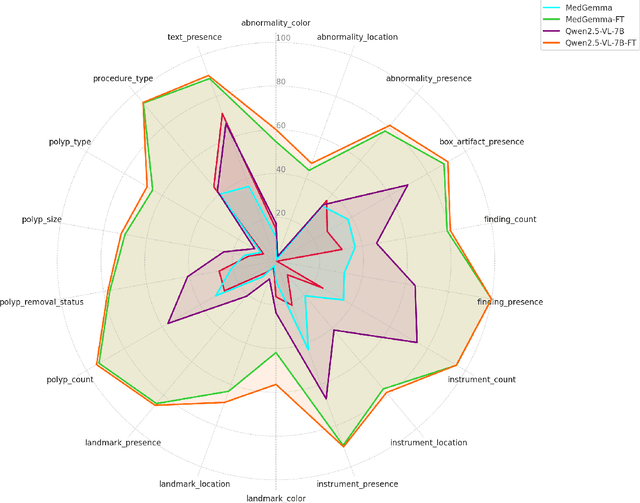
Abstract:Medical Visual Question Answering (MedVQA) is a promising field for developing clinical decision support systems, yet progress is often limited by the available datasets, which can lack clinical complexity and visual diversity. To address these gaps, we introduce Kvasir-VQA-x1, a new, large-scale dataset for gastrointestinal (GI) endoscopy. Our work significantly expands upon the original Kvasir-VQA by incorporating 159,549 new question-answer pairs that are designed to test deeper clinical reasoning. We developed a systematic method using large language models to generate these questions, which are stratified by complexity to better assess a model's inference capabilities. To ensure our dataset prepares models for real-world clinical scenarios, we have also introduced a variety of visual augmentations that mimic common imaging artifacts. The dataset is structured to support two main evaluation tracks: one for standard VQA performance and another to test model robustness against these visual perturbations. By providing a more challenging and clinically relevant benchmark, Kvasir-VQA-x1 aims to accelerate the development of more reliable and effective multimodal AI systems for use in clinical settings. The dataset is fully accessible and adheres to FAIR data principles, making it a valuable resource for the wider research community. Code and data: https://github.com/Simula/Kvasir-VQA-x1 and https://huggingface.co/datasets/SimulaMet/Kvasir-VQA-x1
SoccerChat: Integrating Multimodal Data for Enhanced Soccer Game Understanding
May 22, 2025Abstract:The integration of artificial intelligence in sports analytics has transformed soccer video understanding, enabling real-time, automated insights into complex game dynamics. Traditional approaches rely on isolated data streams, limiting their effectiveness in capturing the full context of a match. To address this, we introduce SoccerChat, a multimodal conversational AI framework that integrates visual and textual data for enhanced soccer video comprehension. Leveraging the extensive SoccerNet dataset, enriched with jersey color annotations and automatic speech recognition (ASR) transcripts, SoccerChat is fine-tuned on a structured video instruction dataset to facilitate accurate game understanding, event classification, and referee decision making. We benchmark SoccerChat on action classification and referee decision-making tasks, demonstrating its performance in general soccer event comprehension while maintaining competitive accuracy in referee decision making. Our findings highlight the importance of multimodal integration in advancing soccer analytics, paving the way for more interactive and explainable AI-driven sports analysis. https://github.com/simula/SoccerChat
Point, Detect, Count: Multi-Task Medical Image Understanding with Instruction-Tuned Vision-Language Models
May 22, 2025Abstract:We investigate fine-tuning Vision-Language Models (VLMs) for multi-task medical image understanding, focusing on detection, localization, and counting of findings in medical images. Our objective is to evaluate whether instruction-tuned VLMs can simultaneously improve these tasks, with the goal of enhancing diagnostic accuracy and efficiency. Using MedMultiPoints, a multimodal dataset with annotations from endoscopy (polyps and instruments) and microscopy (sperm cells), we reformulate each task into instruction-based prompts suitable for vision-language reasoning. We fine-tune Qwen2.5-VL-7B-Instruct using Low-Rank Adaptation (LoRA) across multiple task combinations. Results show that multi-task training improves robustness and accuracy. For example, it reduces the Count Mean Absolute Error (MAE) and increases Matching Accuracy in the Counting + Pointing task. However, trade-offs emerge, such as more zero-case point predictions, indicating reduced reliability in edge cases despite overall performance gains. Our study highlights the potential of adapting general-purpose VLMs to specialized medical tasks via prompt-driven fine-tuning. This approach mirrors clinical workflows, where radiologists simultaneously localize, count, and describe findings - demonstrating how VLMs can learn composite diagnostic reasoning patterns. The model produces interpretable, structured outputs, offering a promising step toward explainable and versatile medical AI. Code, model weights, and scripts will be released for reproducibility at https://github.com/simula/PointDetectCount.
Energy-Conscious LLM Decoding: Impact of Text Generation Strategies on GPU Energy Consumption
Feb 17, 2025



Abstract:Decoding strategies significantly influence the quality and diversity of the generated texts in large language models (LLMs), yet their impact on computational resource consumption, particularly GPU energy usage, is insufficiently studied. This paper investigates the relationship between text generation decoding methods and energy efficiency, focusing on the trade-off between generation quality and GPU energy consumption across diverse tasks and decoding configurations. By benchmarking multiple strategies across different text generation tasks, such as Translation, Code Summarization, and Math Problem Solving, we reveal how selecting appropriate decoding techniques with their tuned hyperparameters affects text quality and has measurable implications for resource utilization, emphasizing the need for balanced optimization. To the best of our knowledge, this study is among the first to explore decoding strategies in LLMs through the lens of energy consumption, offering actionable insights for designing resource-aware applications that maintain high-quality text generation.
Principal Components for Neural Network Initialization
Jan 31, 2025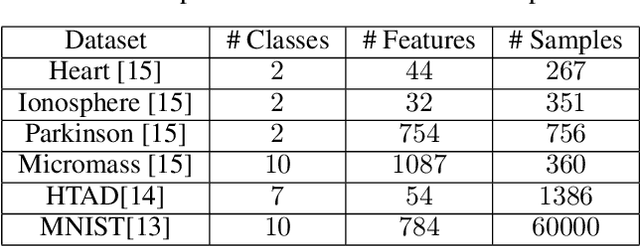
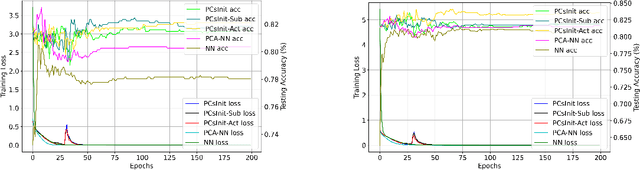
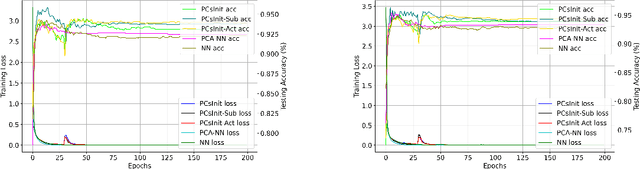
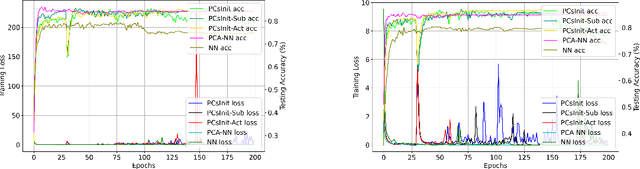
Abstract:Principal Component Analysis (PCA) is a commonly used tool for dimension reduction and denoising. Therefore, it is also widely used on the data prior to training a neural network. However, this approach can complicate the explanation of explainable AI (XAI) methods for the decision of the model. In this work, we analyze the potential issues with this approach and propose Principal Components-based Initialization (PCsInit), a strategy to incorporate PCA into the first layer of a neural network via initialization of the first layer in the network with the principal components, and its two variants PCsInit-Act and PCsInit-Sub. Explanations using these strategies are as direct and straightforward as for neural networks and are simpler than using PCA prior to training a neural network on the principal components. Moreover, as will be illustrated in the experiments, such training strategies can also allow further improvement of training via backpropagation.
DPERC: Direct Parameter Estimation for Mixed Data
Jan 17, 2025
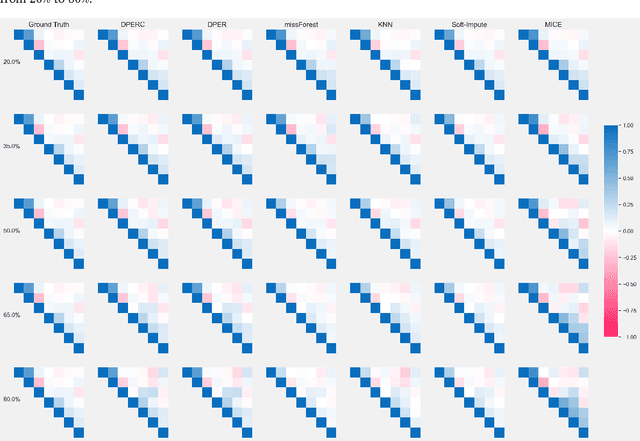
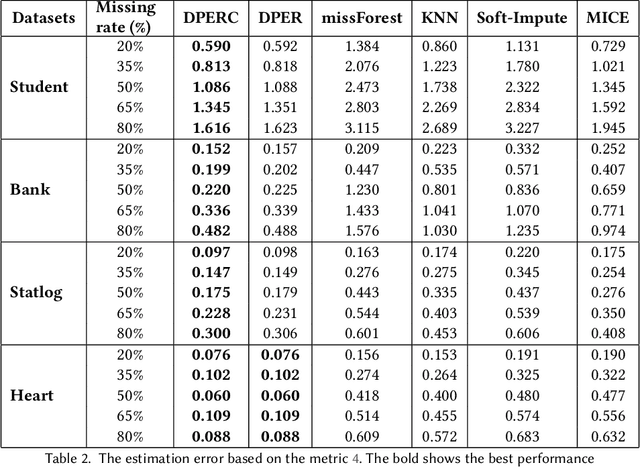
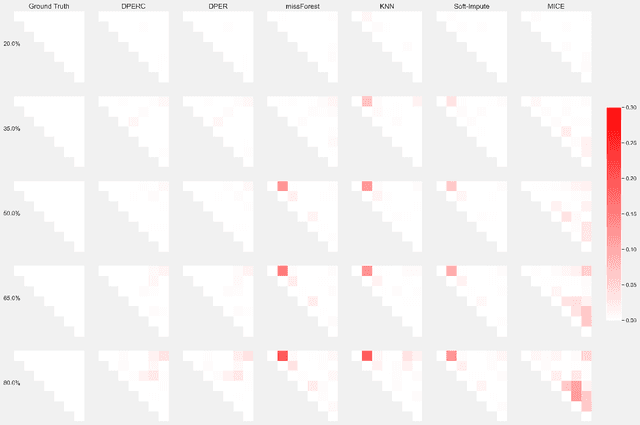
Abstract:The covariance matrix is a foundation in numerous statistical and machine-learning applications such as Principle Component Analysis, Correlation Heatmap, etc. However, missing values within datasets present a formidable obstacle to accurately estimating this matrix. While imputation methods offer one avenue for addressing this challenge, they often entail a trade-off between computational efficiency and estimation accuracy. Consequently, attention has shifted towards direct parameter estimation, given its precision and reduced computational burden. In this paper, we propose Direct Parameter Estimation for Randomly Missing Data with Categorical Features (DPERC), an efficient approach for direct parameter estimation tailored to mixed data that contains missing values within continuous features. Our method is motivated by leveraging information from categorical features, which can significantly enhance covariance matrix estimation for continuous features. Our approach effectively harnesses the information embedded within mixed data structures. Through comprehensive evaluations of diverse datasets, we demonstrate the competitive performance of DPERC compared to various contemporary techniques. In addition, we also show by experiments that DPERC is a valuable tool for visualizing the correlation heatmap.
Missing data imputation for noisy time-series data and applications in healthcare
Dec 15, 2024


Abstract:Healthcare time series data is vital for monitoring patient activity but often contains noise and missing values due to various reasons such as sensor errors or data interruptions. Imputation, i.e., filling in the missing values, is a common way to deal with this issue. In this study, we compare imputation methods, including Multiple Imputation with Random Forest (MICE-RF) and advanced deep learning approaches (SAITS, BRITS, Transformer) for noisy, missing time series data in terms of MAE, F1-score, AUC, and MCC, across missing data rates (10 % - 80 %). Our results show that MICE-RF can effectively impute missing data compared to deep learning methods and the improvement in classification of data imputed indicates that imputation can have denoising effects. Therefore, using an imputation algorithm on time series with missing data can, at the same time, offer denoising effects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge