Pegah Salehi
From Flat to Feeling: A Feasibility and Impact Study on Dynamic Facial Emotions in AI-Generated Avatars
Jun 16, 2025
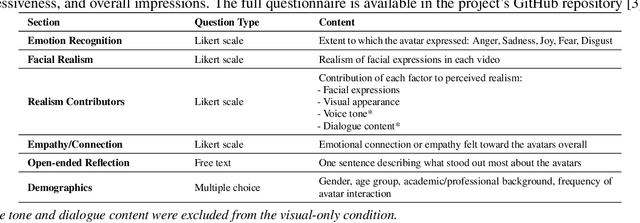
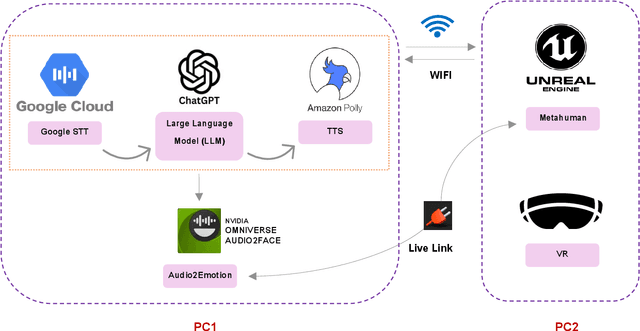

Abstract:Dynamic facial emotion is essential for believable AI-generated avatars; however, most systems remain visually inert, limiting their utility in high-stakes simulations such as virtual training for investigative interviews with abused children. We introduce and evaluate a real-time architecture fusing Unreal Engine 5 MetaHuman rendering with NVIDIA Omniverse Audio2Face to translate vocal prosody into high-fidelity facial expressions on photorealistic child avatars. We implemented a distributed two-PC setup that decouples language processing and speech synthesis from GPU-intensive rendering, designed to support low-latency interaction in desktop and VR environments. A between-subjects study ($N=70$) using audio+visual and visual-only conditions assessed perceptual impacts as participants rated emotional clarity, facial realism, and empathy for two avatars expressing joy, sadness, and anger. Results demonstrate that avatars could express emotions recognizably, with sadness and joy achieving high identification rates. However, anger recognition significantly dropped without audio, highlighting the importance of congruent vocal cues for high-arousal emotions. Interestingly, removing audio boosted perceived facial realism, suggesting that audiovisual desynchrony remains a key design challenge. These findings confirm the technical feasibility of generating emotionally expressive avatars and provide guidance for improving non-verbal communication in sensitive training simulations.
Comparative Analysis of Audio Feature Extraction for Real-Time Talking Portrait Synthesis
Nov 20, 2024



Abstract:This paper examines the integration of real-time talking-head generation for interviewer training, focusing on overcoming challenges in Audio Feature Extraction (AFE), which often introduces latency and limits responsiveness in real-time applications. To address these issues, we propose and implement a fully integrated system that replaces conventional AFE models with Open AI's Whisper, leveraging its encoder to optimize processing and improve overall system efficiency. Our evaluation of two open-source real-time models across three different datasets shows that Whisper not only accelerates processing but also improves specific aspects of rendering quality, resulting in more realistic and responsive talking-head interactions. These advancements make the system a more effective tool for immersive, interactive training applications, expanding the potential of AI-driven avatars in interviewer training.
SinGAN-Seg: Synthetic Training Data Generation for Medical Image Segmentation
Jun 29, 2021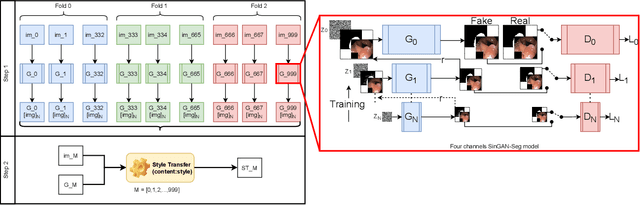
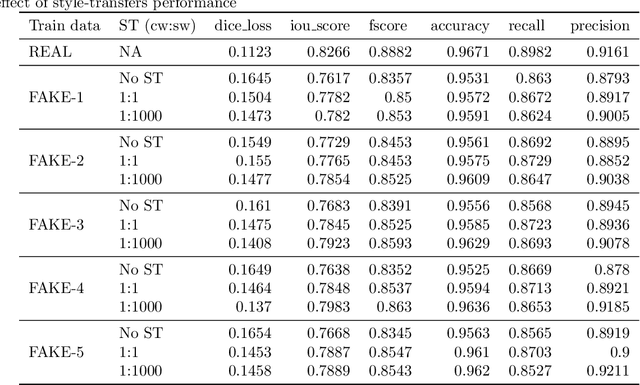
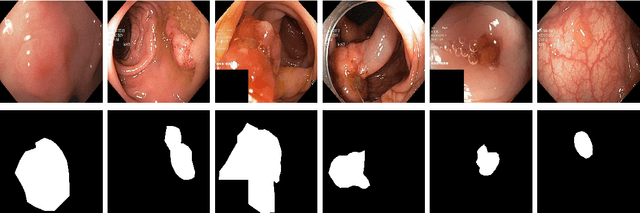
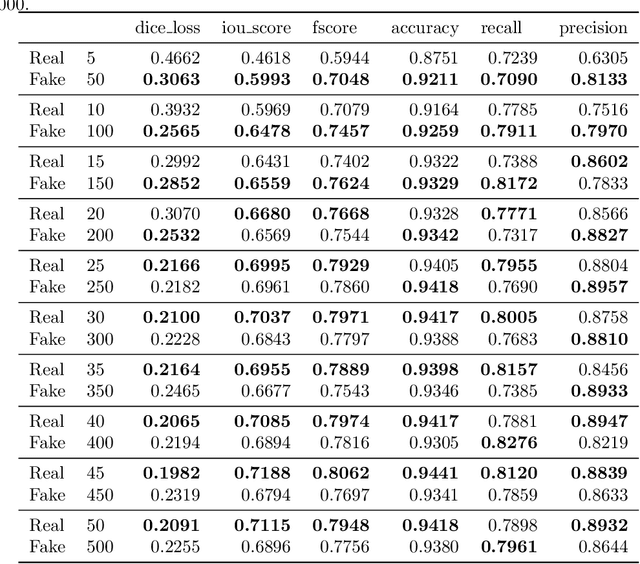
Abstract:Processing medical data to find abnormalities is a time-consuming and costly task, requiring tremendous efforts from medical experts. Therefore, Ai has become a popular tool for the automatic processing of medical data, acting as a supportive tool for doctors. AI tools highly depend on data for training the models. However, there are several constraints to access to large amounts of medical data to train machine learning algorithms in the medical domain, e.g., due to privacy concerns and the costly, time-consuming medical data annotation process. To address this, in this paper we present a novel synthetic data generation pipeline called SinGAN-Seg to produce synthetic medical data with the corresponding annotated ground truth masks. We show that these synthetic data generation pipelines can be used as an alternative to bypass privacy concerns and as an alternative way to produce artificial segmentation datasets with corresponding ground truth masks to avoid the tedious medical data annotation process. As a proof of concept, we used an open polyp segmentation dataset. By training UNet++ using both the real polyp segmentation dataset and the corresponding synthetic dataset generated from the SinGAN-Seg pipeline, we show that the synthetic data can achieve a very close performance to the real data when the real segmentation datasets are large enough. In addition, we show that synthetic data generated from the SinGAN-Seg pipeline improving the performance of segmentation algorithms when the training dataset is very small. Since our SinGAN-Seg pipeline is applicable for any medical dataset, this pipeline can be used with any other segmentation datasets.
Generative Adversarial Networks (GANs): An Overview of Theoretical Model, Evaluation Metrics, and Recent Developments
May 27, 2020



Abstract:One of the most significant challenges in statistical signal processing and machine learning is how to obtain a generative model that can produce samples of large-scale data distribution, such as images and speeches. Generative Adversarial Network (GAN) is an effective method to address this problem. The GANs provide an appropriate way to learn deep representations without widespread use of labeled training data. This approach has attracted the attention of many researchers in computer vision since it can generate a large amount of data without precise modeling of the probability density function (PDF). In GANs, the generative model is estimated via a competitive process where the generator and discriminator networks are trained simultaneously. The generator learns to generate plausible data, and the discriminator learns to distinguish fake data created by the generator from real data samples. Given the rapid growth of GANs over the last few years and their application in various fields, it is necessary to investigate these networks accurately. In this paper, after introducing the main concepts and the theory of GAN, two new deep generative models are compared, the evaluation metrics utilized in the literature and challenges of GANs are also explained. Moreover, the most remarkable GAN architectures are categorized and discussed. Finally, the essential applications in computer vision are examined.
Pix2Pix-based Stain-to-Stain Translation: A Solution for Robust Stain Normalization in Histopathology Images Analysis
Feb 03, 2020



Abstract:The diagnosis of cancer is mainly performed by visual analysis of the pathologists, through examining the morphology of the tissue slices and the spatial arrangement of the cells. If the microscopic image of a specimen is not stained, it will look colorless and textured. Therefore, chemical staining is required to create contrast and help identify specific tissue components. During tissue preparation due to differences in chemicals, scanners, cutting thicknesses, and laboratory protocols, similar tissues are usually varied significantly in appearance. This diversity in staining, in addition to Interpretive disparity among pathologists more is one of the main challenges in designing robust and flexible systems for automated analysis. To address the staining color variations, several methods for normalizing stain have been proposed. In our proposed method, a Stain-to-Stain Translation (STST) approach is used to stain normalization for Hematoxylin and Eosin (H&E) stained histopathology images, which learns not only the specific color distribution but also the preserves corresponding histopathological pattern. We perform the process of translation based on the pix2pix framework, which uses the conditional generator adversarial networks (cGANs). Our approach showed excellent results, both mathematically and experimentally against the state of the art methods. We have made the source code publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge