Binod Bhattarai
Med-MMFL: A Multimodal Federated Learning Benchmark in Healthcare
Feb 04, 2026Abstract:Federated learning (FL) enables collaborative model training across decentralized medical institutions while preserving data privacy. However, medical FL benchmarks remain scarce, with existing efforts focusing mainly on unimodal or bimodal modalities and a limited range of medical tasks. This gap underscores the need for standardized evaluation to advance systematic understanding in medical MultiModal FL (MMFL). To this end, we introduce Med-MMFL, the first comprehensive MMFL benchmark for the medical domain, encompassing diverse modalities, tasks, and federation scenarios. Our benchmark evaluates six representative state-of-the-art FL algorithms, covering different aggregation strategies, loss formulations, and regularization techniques. It spans datasets with 2 to 4 modalities, comprising a total of 10 unique medical modalities, including text, pathology images, ECG, X-ray, radiology reports, and multiple MRI sequences. Experiments are conducted across naturally federated, synthetic IID, and synthetic non-IID settings to simulate real-world heterogeneity. We assess segmentation, classification, modality alignment (retrieval), and VQA tasks. To support reproducibility and fair comparison of future multimodal federated learning (MMFL) methods under realistic medical settings, we release the complete benchmark implementation, including data processing and partitioning pipelines, at https://github.com/bhattarailab/Med-MMFL-Benchmark .
Local K-Similarity Constraint for Federated Learning with Label Noise
Nov 09, 2025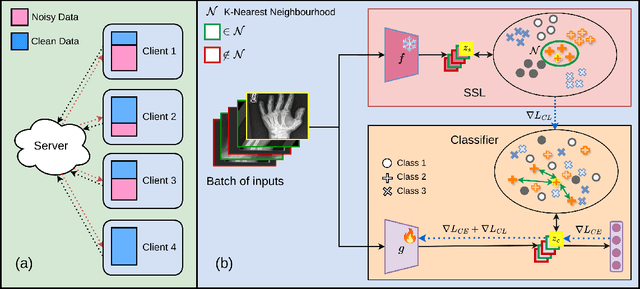
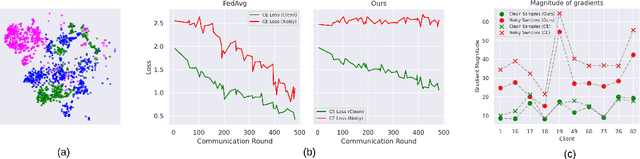
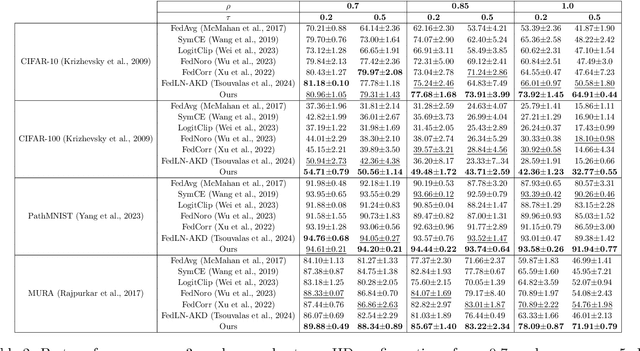
Abstract:Federated learning on clients with noisy labels is a challenging problem, as such clients can infiltrate the global model, impacting the overall generalizability of the system. Existing methods proposed to handle noisy clients assume that a sufficient number of clients with clean labels are available, which can be leveraged to learn a robust global model while dampening the impact of noisy clients. This assumption fails when a high number of heterogeneous clients contain noisy labels, making the existing approaches ineffective. In such scenarios, it is important to locally regularize the clients before communication with the global model, to ensure the global model isn't corrupted by noisy clients. While pre-trained self-supervised models can be effective for local regularization, existing centralized approaches relying on pretrained initialization are impractical in a federated setting due to the potentially large size of these models, which increases communication costs. In that line, we propose a regularization objective for client models that decouples the pre-trained and classification models by enforcing similarity between close data points within the client. We leverage the representation space of a self-supervised pretrained model to evaluate the closeness among examples. This regularization, when applied with the standard objective function for the downstream task in standard noisy federated settings, significantly improves performance, outperforming existing state-of-the-art federated methods in multiple computer vision and medical image classification benchmarks. Unlike other techniques that rely on self-supervised pretrained initialization, our method does not require the pretrained model and classifier backbone to share the same architecture, making it architecture-agnostic.
Estimating 2D Keypoints of Surgical Tools Using Vision-Language Models with Low-Rank Adaptation
Aug 28, 2025



Abstract:This paper presents a novel pipeline for 2D keypoint estima- tion of surgical tools by leveraging Vision Language Models (VLMs) fine- tuned using a low rank adjusting (LoRA) technique. Unlike traditional Convolutional Neural Network (CNN) or Transformer-based approaches, which often suffer from overfitting in small-scale medical datasets, our method harnesses the generalization capabilities of pre-trained VLMs. We carefully design prompts to create an instruction-tuning dataset and use them to align visual features with semantic keypoint descriptions. Experimental results show that with only two epochs of fine tuning, the adapted VLM outperforms the baseline models, demonstrating the ef- fectiveness of LoRA in low-resource scenarios. This approach not only improves keypoint detection performance, but also paves the way for future work in 3D surgical hands and tools pose estimation.
Effect of Data Augmentation on Conformal Prediction for Diabetic Retinopathy
Aug 19, 2025Abstract:The clinical deployment of deep learning models for high-stakes tasks such as diabetic retinopathy (DR) grading requires demonstrable reliability. While models achieve high accuracy, their clinical utility is limited by a lack of robust uncertainty quantification. Conformal prediction (CP) offers a distribution-free framework to generate prediction sets with statistical guarantees of coverage. However, the interaction between standard training practices like data augmentation and the validity of these guarantees is not well understood. In this study, we systematically investigate how different data augmentation strategies affect the performance of conformal predictors for DR grading. Using the DDR dataset, we evaluate two backbone architectures -- ResNet-50 and a Co-Scale Conv-Attentional Transformer (CoaT) -- trained under five augmentation regimes: no augmentation, standard geometric transforms, CLAHE, Mixup, and CutMix. We analyze the downstream effects on conformal metrics, including empirical coverage, average prediction set size, and correct efficiency. Our results demonstrate that sample-mixing strategies like Mixup and CutMix not only improve predictive accuracy but also yield more reliable and efficient uncertainty estimates. Conversely, methods like CLAHE can negatively impact model certainty. These findings highlight the need to co-design augmentation strategies with downstream uncertainty quantification in mind to build genuinely trustworthy AI systems for medical imaging.
Addressing Bias in VLMs for Glaucoma Detection Without Protected Attribute Supervision
Aug 12, 2025Abstract:Vision-Language Models (VLMs) have achieved remarkable success on multimodal tasks such as image-text retrieval and zero-shot classification, yet they can exhibit demographic biases even when explicit protected attributes are absent during training. In this work, we focus on automated glaucoma screening from retinal fundus images, a critical application given that glaucoma is a leading cause of irreversible blindness and disproportionately affects underserved populations. Building on a reweighting-based contrastive learning framework, we introduce an attribute-agnostic debiasing method that (i) infers proxy subgroups via unsupervised clustering of image-image embeddings, (ii) computes gradient-similarity weights between the CLIP-style multimodal loss and a SimCLR-style image-pair contrastive loss, and (iii) applies these weights in a joint, top-$k$ weighted objective to upweight underperforming clusters. This label-free approach adaptively targets the hardest examples, thereby reducing subgroup disparities. We evaluate our method on the Harvard FairVLMed glaucoma subset, reporting Equalized Odds Distance (EOD), Equalized Subgroup AUC (ES AUC), and Groupwise AUC to demonstrate equitable performance across inferred demographic subgroups.
NERO: Explainable Out-of-Distribution Detection with Neuron-level Relevance
Jun 18, 2025Abstract:Ensuring reliability is paramount in deep learning, particularly within the domain of medical imaging, where diagnostic decisions often hinge on model outputs. The capacity to separate out-of-distribution (OOD) samples has proven to be a valuable indicator of a model's reliability in research. In medical imaging, this is especially critical, as identifying OOD inputs can help flag potential anomalies that might otherwise go undetected. While many OOD detection methods rely on feature or logit space representations, recent works suggest these approaches may not fully capture OOD diversity. To address this, we propose a novel OOD scoring mechanism, called NERO, that leverages neuron-level relevance at the feature layer. Specifically, we cluster neuron-level relevance for each in-distribution (ID) class to form representative centroids and introduce a relevance distance metric to quantify a new sample's deviation from these centroids, enhancing OOD separability. Additionally, we refine performance by incorporating scaled relevance in the bias term and combining feature norms. Our framework also enables explainable OOD detection. We validate its effectiveness across multiple deep learning architectures on the gastrointestinal imaging benchmarks Kvasir and GastroVision, achieving improvements over state-of-the-art OOD detection methods.
Federated Foundation Model for GI Endoscopy Images
May 30, 2025Abstract:Gastrointestinal (GI) endoscopy is essential in identifying GI tract abnormalities in order to detect diseases in their early stages and improve patient outcomes. Although deep learning has shown success in supporting GI diagnostics and decision-making, these models require curated datasets with labels that are expensive to acquire. Foundation models offer a promising solution by learning general-purpose representations, which can be finetuned for specific tasks, overcoming data scarcity. Developing foundation models for medical imaging holds significant potential, but the sensitive and protected nature of medical data presents unique challenges. Foundation model training typically requires extensive datasets, and while hospitals generate large volumes of data, privacy restrictions prevent direct data sharing, making foundation model training infeasible in most scenarios. In this work, we propose a FL framework for training foundation models for gastroendoscopy imaging, enabling data to remain within local hospital environments while contributing to a shared model. We explore several established FL algorithms, assessing their suitability for training foundation models without relying on task-specific labels, conducting experiments in both homogeneous and heterogeneous settings. We evaluate the trained foundation model on three critical downstream tasks--classification, detection, and segmentation--and demonstrate that it achieves improved performance across all tasks, highlighting the effectiveness of our approach in a federated, privacy-preserving setting.
Hallucination-Aware Multimodal Benchmark for Gastrointestinal Image Analysis with Large Vision-Language Models
May 11, 2025Abstract:Vision-Language Models (VLMs) are becoming increasingly popular in the medical domain, bridging the gap between medical images and clinical language. Existing VLMs demonstrate an impressive ability to comprehend medical images and text queries to generate detailed, descriptive diagnostic medical reports. However, hallucination--the tendency to generate descriptions that are inconsistent with the visual content--remains a significant issue in VLMs, with particularly severe implications in the medical field. To facilitate VLM research on gastrointestinal (GI) image analysis and study hallucination, we curate a multimodal image-text GI dataset: Gut-VLM. This dataset is created using a two-stage pipeline: first, descriptive medical reports of Kvasir-v2 images are generated using ChatGPT, which introduces some hallucinated or incorrect texts. In the second stage, medical experts systematically review these reports, and identify and correct potential inaccuracies to ensure high-quality, clinically reliable annotations. Unlike traditional datasets that contain only descriptive texts, our dataset also features tags identifying hallucinated sentences and their corresponding corrections. A common approach to reducing hallucination in VLM is to finetune the model on a small-scale, problem-specific dataset. However, we take a different strategy using our dataset. Instead of finetuning the VLM solely for generating textual reports, we finetune it to detect and correct hallucinations, an approach we call hallucination-aware finetuning. Our results show that this approach is better than simply finetuning for descriptive report generation. Additionally, we conduct an extensive evaluation of state-of-the-art VLMs across several metrics, establishing a benchmark. GitHub Repo: https://github.com/bhattarailab/Hallucination-Aware-VLM.
Surgical Vision World Model
Mar 03, 2025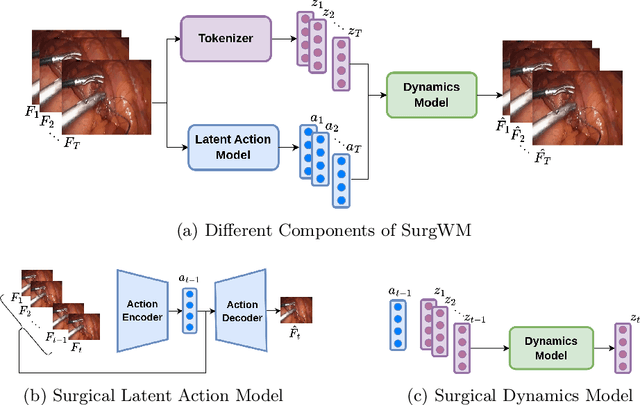
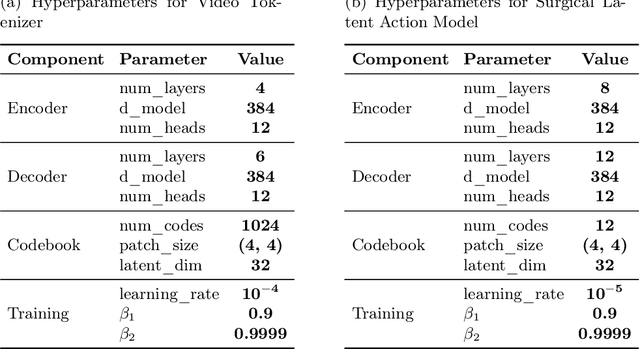
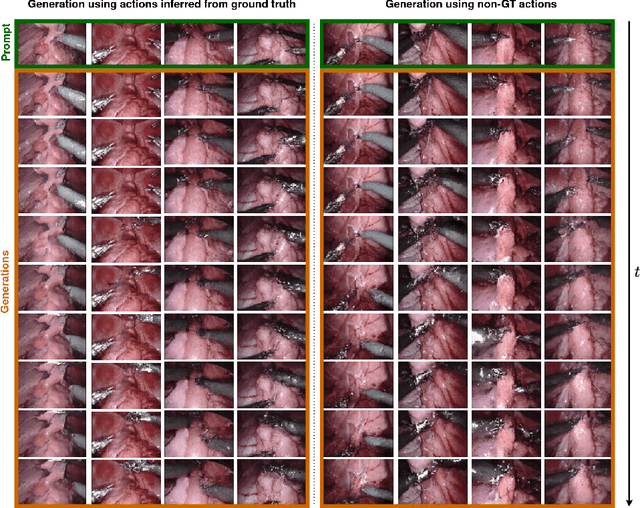
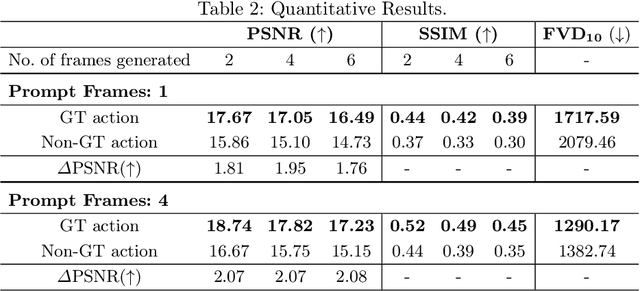
Abstract:Realistic and interactive surgical simulation has the potential to facilitate crucial applications, such as medical professional training and autonomous surgical agent training. In the natural visual domain, world models have enabled action-controlled data generation, demonstrating the potential to train autonomous agents in interactive simulated environments when large-scale real data acquisition is infeasible. However, such works in the surgical domain have been limited to simplified computer simulations, and lack realism. Furthermore, existing literature in world models has predominantly dealt with action-labeled data, limiting their applicability to real-world surgical data, where obtaining action annotation is prohibitively expensive. Inspired by the recent success of Genie in leveraging unlabeled video game data to infer latent actions and enable action-controlled data generation, we propose the first surgical vision world model. The proposed model can generate action-controllable surgical data and the architecture design is verified with extensive experiments on the unlabeled SurgToolLoc-2022 dataset. Codes and implementation details are available at https://github.com/bhattarailab/Surgical-Vision-World-Model
Prompt Augmentation for Self-supervised Text-guided Image Manipulation
Dec 17, 2024
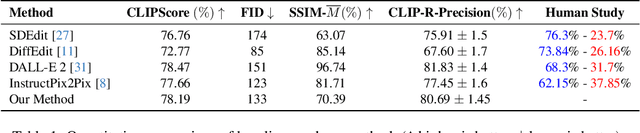
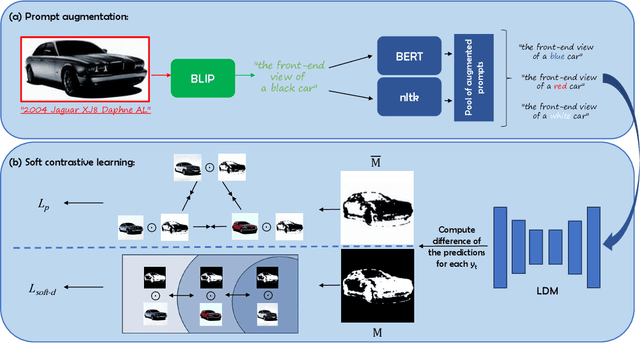
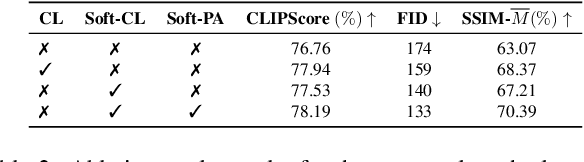
Abstract:Text-guided image editing finds applications in various creative and practical fields. While recent studies in image generation have advanced the field, they often struggle with the dual challenges of coherent image transformation and context preservation. In response, our work introduces prompt augmentation, a method amplifying a single input prompt into several target prompts, strengthening textual context and enabling localised image editing. Specifically, we use the augmented prompts to delineate the intended manipulation area. We propose a Contrastive Loss tailored to driving effective image editing by displacing edited areas and drawing preserved regions closer. Acknowledging the continuous nature of image manipulations, we further refine our approach by incorporating the similarity concept, creating a Soft Contrastive Loss. The new losses are incorporated to the diffusion model, demonstrating improved or competitive image editing results on public datasets and generated images over state-of-the-art approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge