Bidur Khanal
Local K-Similarity Constraint for Federated Learning with Label Noise
Nov 09, 2025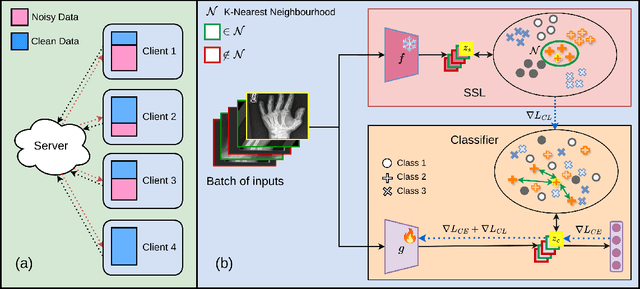
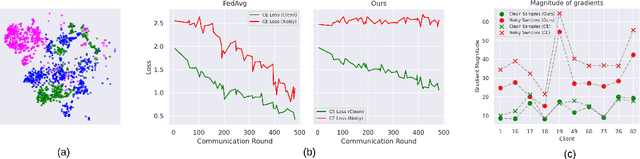
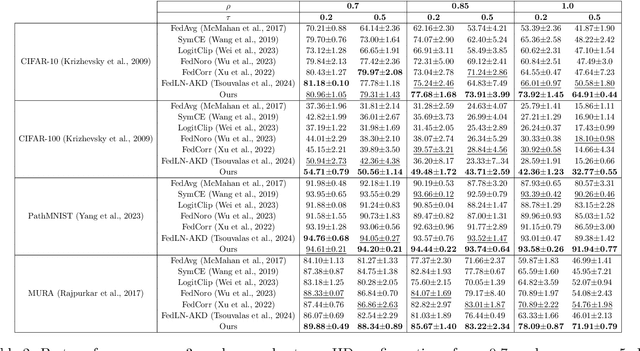
Abstract:Federated learning on clients with noisy labels is a challenging problem, as such clients can infiltrate the global model, impacting the overall generalizability of the system. Existing methods proposed to handle noisy clients assume that a sufficient number of clients with clean labels are available, which can be leveraged to learn a robust global model while dampening the impact of noisy clients. This assumption fails when a high number of heterogeneous clients contain noisy labels, making the existing approaches ineffective. In such scenarios, it is important to locally regularize the clients before communication with the global model, to ensure the global model isn't corrupted by noisy clients. While pre-trained self-supervised models can be effective for local regularization, existing centralized approaches relying on pretrained initialization are impractical in a federated setting due to the potentially large size of these models, which increases communication costs. In that line, we propose a regularization objective for client models that decouples the pre-trained and classification models by enforcing similarity between close data points within the client. We leverage the representation space of a self-supervised pretrained model to evaluate the closeness among examples. This regularization, when applied with the standard objective function for the downstream task in standard noisy federated settings, significantly improves performance, outperforming existing state-of-the-art federated methods in multiple computer vision and medical image classification benchmarks. Unlike other techniques that rely on self-supervised pretrained initialization, our method does not require the pretrained model and classifier backbone to share the same architecture, making it architecture-agnostic.
Hallucination-Aware Multimodal Benchmark for Gastrointestinal Image Analysis with Large Vision-Language Models
May 11, 2025Abstract:Vision-Language Models (VLMs) are becoming increasingly popular in the medical domain, bridging the gap between medical images and clinical language. Existing VLMs demonstrate an impressive ability to comprehend medical images and text queries to generate detailed, descriptive diagnostic medical reports. However, hallucination--the tendency to generate descriptions that are inconsistent with the visual content--remains a significant issue in VLMs, with particularly severe implications in the medical field. To facilitate VLM research on gastrointestinal (GI) image analysis and study hallucination, we curate a multimodal image-text GI dataset: Gut-VLM. This dataset is created using a two-stage pipeline: first, descriptive medical reports of Kvasir-v2 images are generated using ChatGPT, which introduces some hallucinated or incorrect texts. In the second stage, medical experts systematically review these reports, and identify and correct potential inaccuracies to ensure high-quality, clinically reliable annotations. Unlike traditional datasets that contain only descriptive texts, our dataset also features tags identifying hallucinated sentences and their corresponding corrections. A common approach to reducing hallucination in VLM is to finetune the model on a small-scale, problem-specific dataset. However, we take a different strategy using our dataset. Instead of finetuning the VLM solely for generating textual reports, we finetune it to detect and correct hallucinations, an approach we call hallucination-aware finetuning. Our results show that this approach is better than simply finetuning for descriptive report generation. Additionally, we conduct an extensive evaluation of state-of-the-art VLMs across several metrics, establishing a benchmark. GitHub Repo: https://github.com/bhattarailab/Hallucination-Aware-VLM.
Assessing the Performance of the DINOv2 Self-supervised Learning Vision Transformer Model for the Segmentation of the Left Atrium from MRI Images
Nov 14, 2024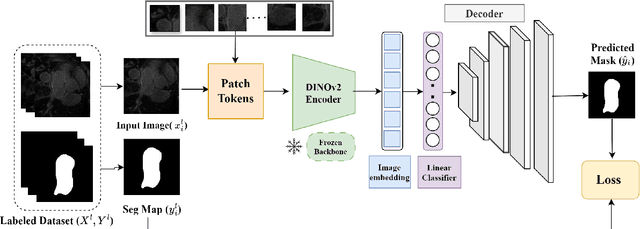
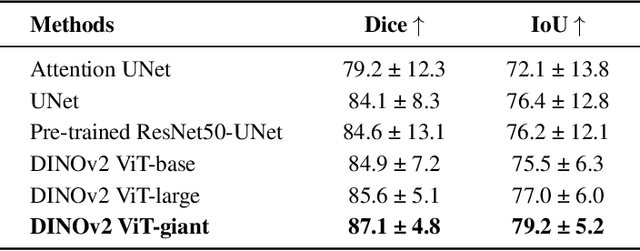
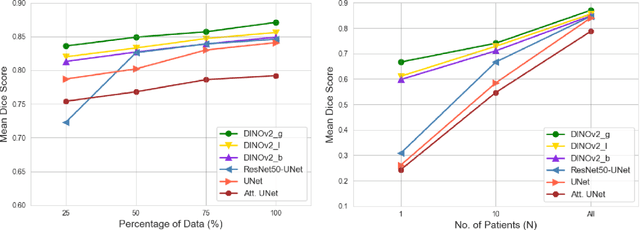
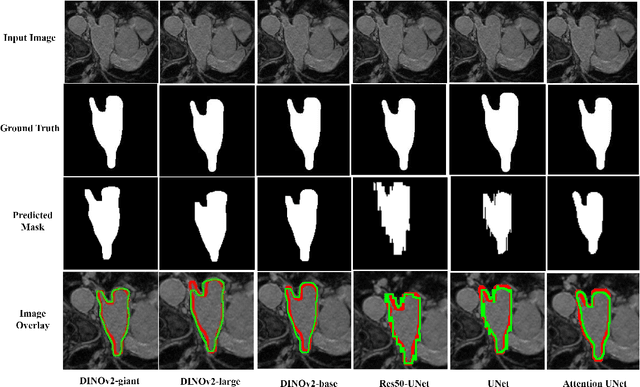
Abstract:Accurate left atrium (LA) segmentation from pre-operative scans is crucial for diagnosing atrial fibrillation, treatment planning, and supporting surgical interventions. While deep learning models are key in medical image segmentation, they often require extensive manually annotated data. Foundation models trained on larger datasets have reduced this dependency, enhancing generalizability and robustness through transfer learning. We explore DINOv2, a self-supervised learning vision transformer trained on natural images, for LA segmentation using MRI. The challenges for LA's complex anatomy, thin boundaries, and limited annotated data make accurate segmentation difficult before & during the image-guided intervention. We demonstrate DINOv2's ability to provide accurate & consistent segmentation, achieving a mean Dice score of .871 & a Jaccard Index of .792 for end-to-end fine-tuning. Through few-shot learning across various data sizes & patient counts, DINOv2 consistently outperforms baseline models. These results suggest that DINOv2 effectively adapts to MRI with limited data, highlighting its potential as a competitive tool for segmentation & encouraging broader use in medical imaging.
Active Label Refinement for Robust Training of Imbalanced Medical Image Classification Tasks in the Presence of High Label Noise
Jul 08, 2024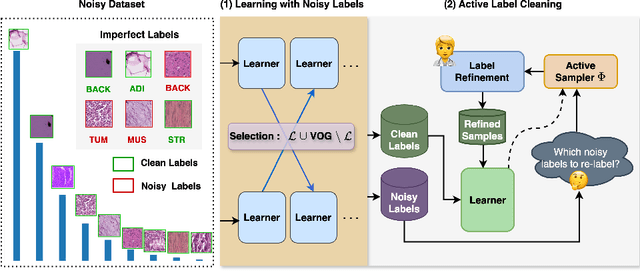

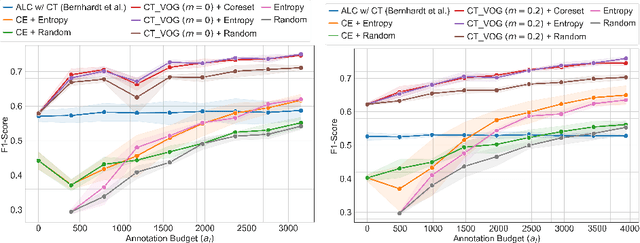
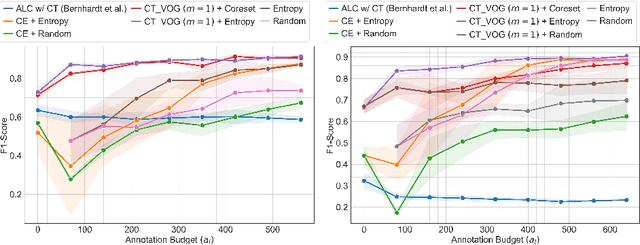
Abstract:The robustness of supervised deep learning-based medical image classification is significantly undermined by label noise. Although several methods have been proposed to enhance classification performance in the presence of noisy labels, they face some challenges: 1) a struggle with class-imbalanced datasets, leading to the frequent overlooking of minority classes as noisy samples; 2) a singular focus on maximizing performance using noisy datasets, without incorporating experts-in-the-loop for actively cleaning the noisy labels. To mitigate these challenges, we propose a two-phase approach that combines Learning with Noisy Labels (LNL) and active learning. This approach not only improves the robustness of medical image classification in the presence of noisy labels, but also iteratively improves the quality of the dataset by relabeling the important incorrect labels, under a limited annotation budget. Furthermore, we introduce a novel Variance of Gradients approach in LNL phase, which complements the loss-based sample selection by also sampling under-represented samples. Using two imbalanced noisy medical classification datasets, we demonstrate that that our proposed technique is superior to its predecessors at handling class imbalance by not misidentifying clean samples from minority classes as mostly noisy samples.
Investigating the Robustness of Vision Transformers against Label Noise in Medical Image Classification
Feb 26, 2024Abstract:Label noise in medical image classification datasets significantly hampers the training of supervised deep learning methods, undermining their generalizability. The test performance of a model tends to decrease as the label noise rate increases. Over recent years, several methods have been proposed to mitigate the impact of label noise in medical image classification and enhance the robustness of the model. Predominantly, these works have employed CNN-based architectures as the backbone of their classifiers for feature extraction. However, in recent years, Vision Transformer (ViT)-based backbones have replaced CNNs, demonstrating improved performance and a greater ability to learn more generalizable features, especially when the dataset is large. Nevertheless, no prior work has rigorously investigated how transformer-based backbones handle the impact of label noise in medical image classification. In this paper, we investigate the architectural robustness of ViT against label noise and compare it to that of CNNs. We use two medical image classification datasets -- COVID-DU-Ex, and NCT-CRC-HE-100K -- both corrupted by injecting label noise at various rates. Additionally, we show that pretraining is crucial for ensuring ViT's improved robustness against label noise in supervised training.
How does self-supervised pretraining improve robustness against noisy labels across various medical image classification datasets?
Jan 15, 2024Abstract:Noisy labels can significantly impact medical image classification, particularly in deep learning, by corrupting learned features. Self-supervised pretraining, which doesn't rely on labeled data, can enhance robustness against noisy labels. However, this robustness varies based on factors like the number of classes, dataset complexity, and training size. In medical images, subtle inter-class differences and modality-specific characteristics add complexity. Previous research hasn't comprehensively explored the interplay between self-supervised learning and robustness against noisy labels in medical image classification, considering all these factors. In this study, we address three key questions: i) How does label noise impact various medical image classification datasets? ii) Which types of medical image datasets are more challenging to learn and more affected by label noise? iii) How do different self-supervised pretraining methods enhance robustness across various medical image datasets? Our results show that DermNet, among five datasets (Fetal plane, DermNet, COVID-DU-Ex, MURA, NCT-CRC-HE-100K), is the most challenging but exhibits greater robustness against noisy labels. Additionally, contrastive learning stands out among the eight self-supervised methods as the most effective approach to enhance robustness against noisy labels.
Medical Vision Language Pretraining: A survey
Dec 11, 2023Abstract:Medical Vision Language Pretraining (VLP) has recently emerged as a promising solution to the scarcity of labeled data in the medical domain. By leveraging paired/unpaired vision and text datasets through self-supervised learning, models can be trained to acquire vast knowledge and learn robust feature representations. Such pretrained models have the potential to enhance multiple downstream medical tasks simultaneously, reducing the dependency on labeled data. However, despite recent progress and its potential, there is no such comprehensive survey paper that has explored the various aspects and advancements in medical VLP. In this paper, we specifically review existing works through the lens of different pretraining objectives, architectures, downstream evaluation tasks, and datasets utilized for pretraining and downstream tasks. Subsequently, we delve into current challenges in medical VLP, discussing existing and potential solutions, and conclude by highlighting future directions. To the best of our knowledge, this is the first survey focused on medical VLP.
Improving Medical Image Classification in Noisy Labels Using Only Self-supervised Pretraining
Aug 08, 2023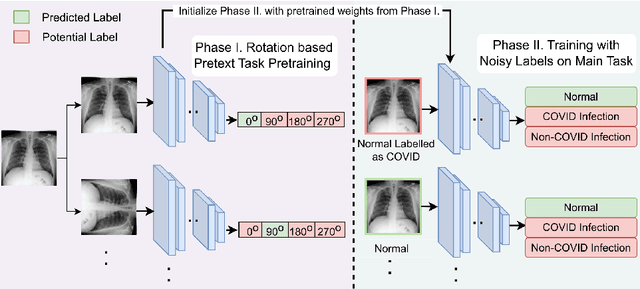

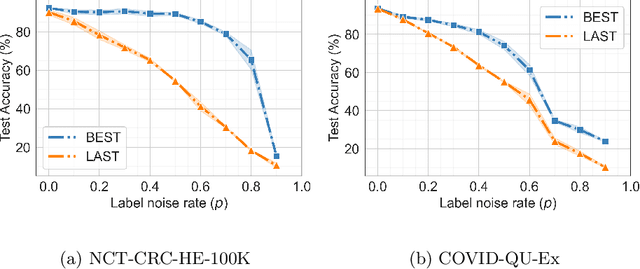
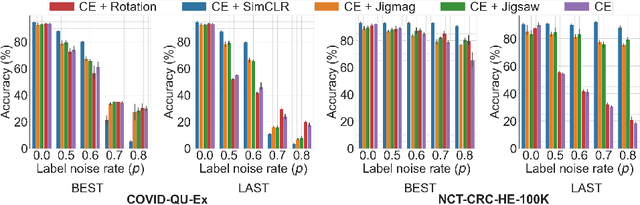
Abstract:Noisy labels hurt deep learning-based supervised image classification performance as the models may overfit the noise and learn corrupted feature extractors. For natural image classification training with noisy labeled data, model initialization with contrastive self-supervised pretrained weights has shown to reduce feature corruption and improve classification performance. However, no works have explored: i) how other self-supervised approaches, such as pretext task-based pretraining, impact the learning with noisy label, and ii) any self-supervised pretraining methods alone for medical images in noisy label settings. Medical images often feature smaller datasets and subtle inter class variations, requiring human expertise to ensure correct classification. Thus, it is not clear if the methods improving learning with noisy labels in natural image datasets such as CIFAR would also help with medical images. In this work, we explore contrastive and pretext task-based self-supervised pretraining to initialize the weights of a deep learning classification model for two medical datasets with self-induced noisy labels -- NCT-CRC-HE-100K tissue histological images and COVID-QU-Ex chest X-ray images. Our results show that models initialized with pretrained weights obtained from self-supervised learning can effectively learn better features and improve robustness against noisy labels.
M-VAAL: Multimodal Variational Adversarial Active Learning for Downstream Medical Image Analysis Tasks
Jun 21, 2023Abstract:Acquiring properly annotated data is expensive in the medical field as it requires experts, time-consuming protocols, and rigorous validation. Active learning attempts to minimize the need for large annotated samples by actively sampling the most informative examples for annotation. These examples contribute significantly to improving the performance of supervised machine learning models, and thus, active learning can play an essential role in selecting the most appropriate information in deep learning-based diagnosis, clinical assessments, and treatment planning. Although some existing works have proposed methods for sampling the best examples for annotation in medical image analysis, they are not task-agnostic and do not use multimodal auxiliary information in the sampler, which has the potential to increase robustness. Therefore, in this work, we propose a Multimodal Variational Adversarial Active Learning (M-VAAL) method that uses auxiliary information from additional modalities to enhance the active sampling. We applied our method to two datasets: i) brain tumor segmentation and multi-label classification using the BraTS2018 dataset, and ii) chest X-ray image classification using the COVID-QU-Ex dataset. Our results show a promising direction toward data-efficient learning under limited annotations.
How Does Heterogeneous Label Noise Impact Generalization in Neural Nets?
Jun 29, 2021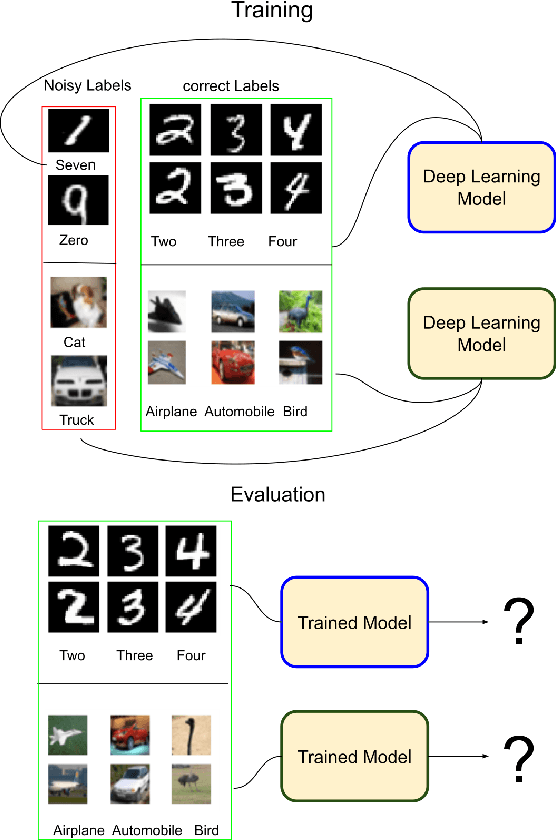
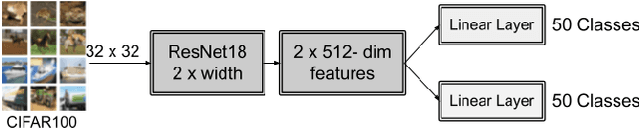
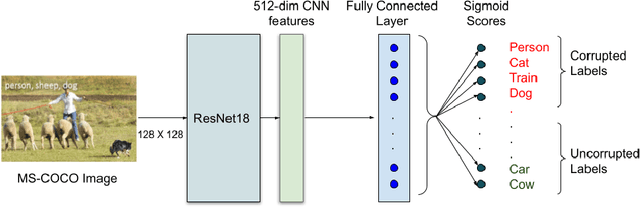
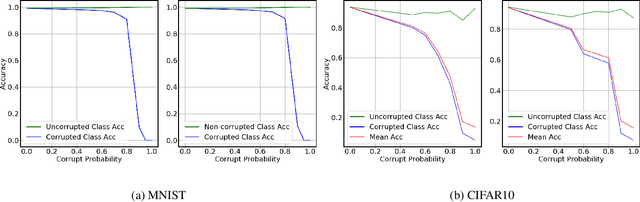
Abstract:Incorrectly labeled examples, or label noise, is common in real-world computer vision datasets. While the impact of label noise on learning in deep neural networks has been studied in prior work, these studies have exclusively focused on homogeneous label noise, i.e., the degree of label noise is the same across all categories. However, in the real-world, label noise is often heterogeneous, with some categories being affected to a greater extent than others. Here, we address this gap in the literature. We hypothesized that heterogeneous label noise would only affect the classes that had label noise unless there was transfer from those classes to the classes without label noise. To test this hypothesis, we designed a series of computer vision studies using MNIST, CIFAR-10, CIFAR-100, and MS-COCO where we imposed heterogeneous label noise during the training of multi-class, multi-task, and multi-label systems. Our results provide evidence in support of our hypothesis: label noise only affects the class affected by it unless there is transfer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge