Bishesh Khanal
NAAMII, Nepal, Tangible Careers
The AI Pyramid A Conceptual Framework for Workforce Capability in the Age of AI
Jan 10, 2026Abstract:Artificial intelligence (AI) represents a qualitative shift in technological change by extending cognitive labor itself rather than merely automating routine tasks. Recent evidence shows that generative AI disproportionately affects highly educated, white collar work, challenging existing assumptions about workforce vulnerability and rendering traditional approaches to digital or AI literacy insufficient. This paper introduces the concept of AI Nativity, the capacity to integrate AI fluidly into everyday reasoning, problem solving, and decision making, and proposes the AI Pyramid, a conceptual framework for organizing human capability in an AI mediated economy. The framework distinguishes three interdependent capability layers: AI Native capability as a universal baseline for participation in AI augmented environments; AI Foundation capability for building, integrating, and sustaining AI enabled systems; and AI Deep capability for advancing frontier AI knowledge and applications. Crucially, the pyramid is not a career ladder but a system level distribution of capabilities required at scale. Building on this structure, the paper argues that effective AI workforce development requires treating capability formation as infrastructure rather than episodic training, centered on problem based learning embedded in work contexts and supported by dynamic skill ontologies and competency based measurement. The framework has implications for organizations, education systems, and governments seeking to align learning, measurement, and policy with the evolving demands of AI mediated work, while addressing productivity, resilience, and inequality at societal scale.
From Development to Deployment of AI-assisted Telehealth and Screening for Vision- and Hearing-threatening diseases in resource-constrained settings: Field Observations, Challenges and Way Forward
Sep 19, 2025


Abstract:Vision- and hearing-threatening diseases cause preventable disability, especially in resource-constrained settings(RCS) with few specialists and limited screening setup. Large scale AI-assisted screening and telehealth has potential to expand early detection, but practical deployment is challenging in paper-based workflows and limited documented field experience exist to build upon. We provide insights on challenges and ways forward in development to adoption of scalable AI-assisted Telehealth and screening in such settings. Specifically, we find that iterative, interdisciplinary collaboration through early prototyping, shadow deployment and continuous feedback is important to build shared understanding as well as reduce usability hurdles when transitioning from paper-based to AI-ready workflows. We find public datasets and AI models highly useful despite poor performance due to domain shift. In addition, we find the need for automated AI-based image quality check to capture gradable images for robust screening in high-volume camps. Our field learning stress the importance of treating AI development and workflow digitization as an end-to-end, iterative co-design process. By documenting these practical challenges and lessons learned, we aim to address the gap in contextual, actionable field knowledge for building real-world AI-assisted telehealth and mass-screening programs in RCS.
Parameter-efficient Fine-tuning for improved Convolutional Baseline for Brain Tumor Segmentation in Sub-Saharan Africa Adult Glioma Dataset
Dec 18, 2024

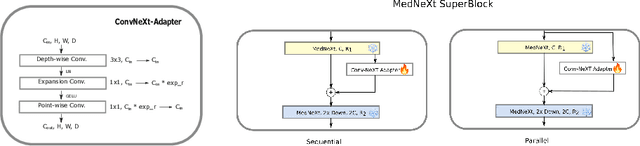
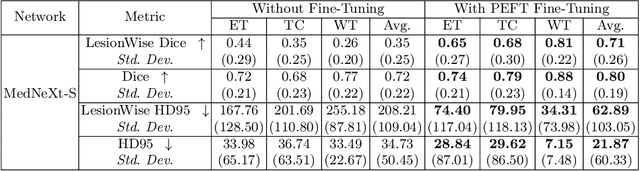
Abstract:Automating brain tumor segmentation using deep learning methods is an ongoing challenge in medical imaging. Multiple lingering issues exist including domain-shift and applications in low-resource settings which brings a unique set of challenges including scarcity of data. As a step towards solving these specific problems, we propose Convolutional adapter-inspired Parameter-efficient Fine-tuning (PEFT) of MedNeXt architecture. To validate our idea, we show our method performs comparable to full fine-tuning with the added benefit of reduced training compute using BraTS-2021 as pre-training dataset and BraTS-Africa as the fine-tuning dataset. BraTS-Africa consists of a small dataset (60 train / 35 validation) from the Sub-Saharan African population with marked shift in the MRI quality compared to BraTS-2021 (1251 train samples). We first show that models trained on BraTS-2021 dataset do not generalize well to BraTS-Africa as shown by 20% reduction in mean dice on BraTS-Africa validation samples. Then, we show that PEFT can leverage both the BraTS-2021 and BraTS-Africa dataset to obtain mean dice of 0.8 compared to 0.72 when trained only on BraTS-Africa. Finally, We show that PEFT (0.80 mean dice) results in comparable performance to full fine-tuning (0.77 mean dice) which may show PEFT to be better on average but the boxplots show that full finetuning results is much lesser variance in performance. Nevertheless, on disaggregation of the dice metrics, we find that the model has tendency to oversegment as shown by high specificity (0.99) compared to relatively low sensitivity(0.75). The source code is available at https://github.com/CAMERA-MRI/SPARK2024/tree/main/PEFT_MedNeXt
NLPineers@ NLU of Devanagari Script Languages 2025: Hate Speech Detection using Ensembling of BERT-based models
Dec 12, 2024



Abstract:This paper explores hate speech detection in Devanagari-scripted languages, focusing on Hindi and Nepali, for Subtask B of the CHIPSAL@COLING 2025 Shared Task. Using a range of transformer-based models such as XLM-RoBERTa, MURIL, and IndicBERT, we examine their effectiveness in navigating the nuanced boundary between hate speech and free expression. Our best performing model, implemented as ensemble of multilingual BERT models achieve Recall of 0.7762 (Rank 3/31 in terms of recall) and F1 score of 0.6914 (Rank 17/31). To address class imbalance, we used backtranslation for data augmentation, and cosine similarity to preserve label consistency after augmentation. This work emphasizes the need for hate speech detection in Devanagari-scripted languages and presents a foundation for further research.
TuneVLSeg: Prompt Tuning Benchmark for Vision-Language Segmentation Models
Oct 07, 2024



Abstract:Vision-Language Models (VLMs) have shown impressive performance in vision tasks, but adapting them to new domains often requires expensive fine-tuning. Prompt tuning techniques, including textual, visual, and multimodal prompting, offer efficient alternatives by leveraging learnable prompts. However, their application to Vision-Language Segmentation Models (VLSMs) and evaluation under significant domain shifts remain unexplored. This work presents an open-source benchmarking framework, TuneVLSeg, to integrate various unimodal and multimodal prompt tuning techniques into VLSMs, making prompt tuning usable for downstream segmentation datasets with any number of classes. TuneVLSeg includes $6$ prompt tuning strategies on various prompt depths used in $2$ VLSMs totaling of $8$ different combinations. We test various prompt tuning on $8$ diverse medical datasets, including $3$ radiology datasets (breast tumor, echocardiograph, chest X-ray pathologies) and $5$ non-radiology datasets (polyp, ulcer, skin cancer), and two natural domain segmentation datasets. Our study found that textual prompt tuning struggles under significant domain shifts, from natural-domain images to medical data. Furthermore, visual prompt tuning, with fewer hyperparameters than multimodal prompt tuning, often achieves performance competitive to multimodal approaches, making it a valuable first attempt. Our work advances the understanding and applicability of different prompt-tuning techniques for robust domain-specific segmentation. The source code is available at https://github.com/naamiinepal/tunevlseg.
VLSM-Adapter: Finetuning Vision-Language Segmentation Efficiently with Lightweight Blocks
May 10, 2024Abstract:Foundation Vision-Language Models (VLMs) trained using large-scale open-domain images and text pairs have recently been adapted to develop Vision-Language Segmentation Models (VLSMs) that allow providing text prompts during inference to guide image segmentation. If robust and powerful VLSMs can be built for medical images, it could aid medical professionals in many clinical tasks where they must spend substantial time delineating the target structure of interest. VLSMs for medical images resort to fine-tuning base VLM or VLSM pretrained on open-domain natural image datasets due to fewer annotated medical image datasets; this fine-tuning is resource-consuming and expensive as it usually requires updating all or a significant fraction of the pretrained parameters. Recently, lightweight blocks called adapters have been proposed in VLMs that keep the pretrained model frozen and only train adapters during fine-tuning, substantially reducing the computing resources required. We introduce a novel adapter, VLSM-Adapter, that can fine-tune pretrained vision-language segmentation models using transformer encoders. Our experiments in widely used CLIP-based segmentation models show that with only 3 million trainable parameters, the VLSM-Adapter outperforms state-of-the-art and is comparable to the upper bound end-to-end fine-tuning. The source code is available at: https://github.com/naamiinepal/vlsm-adapter.
AI-Assisted Cervical Cancer Screening
Mar 18, 2024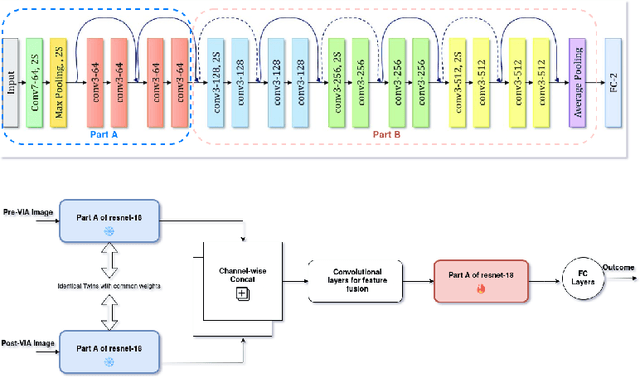

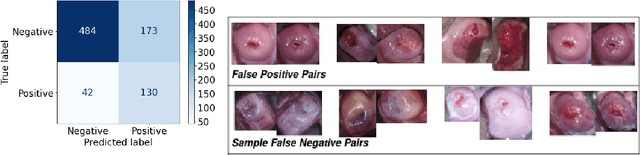
Abstract:Visual Inspection with Acetic Acid (VIA) remains the most feasible cervical cancer screening test in resource-constrained settings of low- and middle-income countries (LMICs), which are often performed screening camps or primary/community health centers by nurses instead of the preferred but unavailable expert Gynecologist. To address the highly subjective nature of the test, various handheld devices integrating cameras or smartphones have been recently explored to capture cervical images during VIA and aid decision-making via telemedicine or AI models. Most studies proposing AI models retrospectively use a relatively small number of already collected images from specific devices, digital cameras, or smartphones; the challenges and protocol for quality image acquisition during VIA in resource-constrained camp settings, challenges in getting gold standard, data imbalance, etc. are often overlooked. We present a novel approach and describe the end-to-end design process to build a robust smartphone-based AI-assisted system that does not require buying a separate integrated device: the proposed protocol for quality image acquisition in resource-constrained settings, dataset collected from 1,430 women during VIA performed by nurses in screening camps, preprocessing pipeline, and training and evaluation of a deep-learning-based classification model aimed to identify (pre)cancerous lesions. Our work shows that the readily available smartphones and a suitable protocol can capture the cervix images with the required details for the VIA test well; the deep-learning-based classification model provides promising results to assist nurses in VIA screening; and provides a direction for large-scale data collection and validation in resource-constrained settings.
Investigating the Robustness of Vision Transformers against Label Noise in Medical Image Classification
Feb 26, 2024Abstract:Label noise in medical image classification datasets significantly hampers the training of supervised deep learning methods, undermining their generalizability. The test performance of a model tends to decrease as the label noise rate increases. Over recent years, several methods have been proposed to mitigate the impact of label noise in medical image classification and enhance the robustness of the model. Predominantly, these works have employed CNN-based architectures as the backbone of their classifiers for feature extraction. However, in recent years, Vision Transformer (ViT)-based backbones have replaced CNNs, demonstrating improved performance and a greater ability to learn more generalizable features, especially when the dataset is large. Nevertheless, no prior work has rigorously investigated how transformer-based backbones handle the impact of label noise in medical image classification. In this paper, we investigate the architectural robustness of ViT against label noise and compare it to that of CNNs. We use two medical image classification datasets -- COVID-DU-Ex, and NCT-CRC-HE-100K -- both corrupted by injecting label noise at various rates. Additionally, we show that pretraining is crucial for ensuring ViT's improved robustness against label noise in supervised training.
How does self-supervised pretraining improve robustness against noisy labels across various medical image classification datasets?
Jan 15, 2024Abstract:Noisy labels can significantly impact medical image classification, particularly in deep learning, by corrupting learned features. Self-supervised pretraining, which doesn't rely on labeled data, can enhance robustness against noisy labels. However, this robustness varies based on factors like the number of classes, dataset complexity, and training size. In medical images, subtle inter-class differences and modality-specific characteristics add complexity. Previous research hasn't comprehensively explored the interplay between self-supervised learning and robustness against noisy labels in medical image classification, considering all these factors. In this study, we address three key questions: i) How does label noise impact various medical image classification datasets? ii) Which types of medical image datasets are more challenging to learn and more affected by label noise? iii) How do different self-supervised pretraining methods enhance robustness across various medical image datasets? Our results show that DermNet, among five datasets (Fetal plane, DermNet, COVID-DU-Ex, MURA, NCT-CRC-HE-100K), is the most challenging but exhibits greater robustness against noisy labels. Additionally, contrastive learning stands out among the eight self-supervised methods as the most effective approach to enhance robustness against noisy labels.
Benchmarking Encoder-Decoder Architectures for Biplanar X-ray to 3D Shape Reconstruction
Sep 26, 2023



Abstract:Various deep learning models have been proposed for 3D bone shape reconstruction from two orthogonal (biplanar) X-ray images. However, it is unclear how these models compare against each other since they are evaluated on different anatomy, cohort and (often privately held) datasets. Moreover, the impact of the commonly optimized image-based segmentation metrics such as dice score on the estimation of clinical parameters relevant in 2D-3D bone shape reconstruction is not well known. To move closer toward clinical translation, we propose a benchmarking framework that evaluates tasks relevant to real-world clinical scenarios, including reconstruction of fractured bones, bones with implants, robustness to population shift, and error in estimating clinical parameters. Our open-source platform provides reference implementations of 8 models (many of whose implementations were not publicly available), APIs to easily collect and preprocess 6 public datasets, and the implementation of automatic clinical parameter and landmark extraction methods. We present an extensive evaluation of 8 2D-3D models on equal footing using 6 public datasets comprising images for four different anatomies. Our results show that attention-based methods that capture global spatial relationships tend to perform better across all anatomies and datasets; performance on clinically relevant subgroups may be overestimated without disaggregated reporting; ribs are substantially more difficult to reconstruct compared to femur, hip and spine; and the dice score improvement does not always bring a corresponding improvement in the automatic estimation of clinically relevant parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge