Kanchan Poudel
AI-Assisted Cervical Cancer Screening
Mar 18, 2024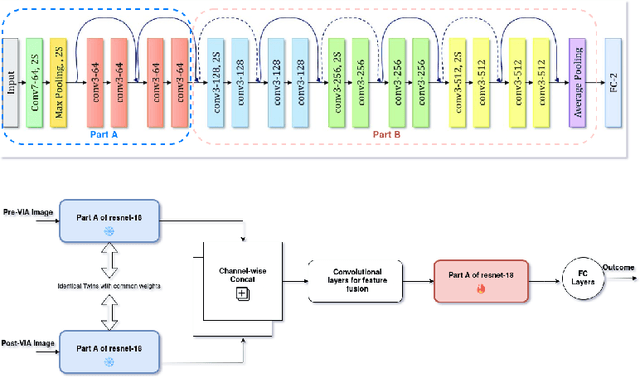

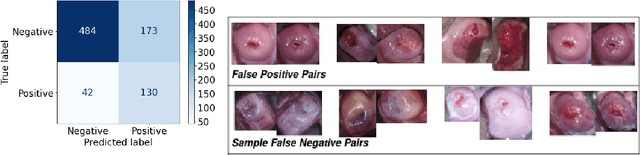
Abstract:Visual Inspection with Acetic Acid (VIA) remains the most feasible cervical cancer screening test in resource-constrained settings of low- and middle-income countries (LMICs), which are often performed screening camps or primary/community health centers by nurses instead of the preferred but unavailable expert Gynecologist. To address the highly subjective nature of the test, various handheld devices integrating cameras or smartphones have been recently explored to capture cervical images during VIA and aid decision-making via telemedicine or AI models. Most studies proposing AI models retrospectively use a relatively small number of already collected images from specific devices, digital cameras, or smartphones; the challenges and protocol for quality image acquisition during VIA in resource-constrained camp settings, challenges in getting gold standard, data imbalance, etc. are often overlooked. We present a novel approach and describe the end-to-end design process to build a robust smartphone-based AI-assisted system that does not require buying a separate integrated device: the proposed protocol for quality image acquisition in resource-constrained settings, dataset collected from 1,430 women during VIA performed by nurses in screening camps, preprocessing pipeline, and training and evaluation of a deep-learning-based classification model aimed to identify (pre)cancerous lesions. Our work shows that the readily available smartphones and a suitable protocol can capture the cervix images with the required details for the VIA test well; the deep-learning-based classification model provides promising results to assist nurses in VIA screening; and provides a direction for large-scale data collection and validation in resource-constrained settings.
Synthetic Boost: Leveraging Synthetic Data for Enhanced Vision-Language Segmentation in Echocardiography
Sep 22, 2023Abstract:Accurate segmentation is essential for echocardiography-based assessment of cardiovascular diseases (CVDs). However, the variability among sonographers and the inherent challenges of ultrasound images hinder precise segmentation. By leveraging the joint representation of image and text modalities, Vision-Language Segmentation Models (VLSMs) can incorporate rich contextual information, potentially aiding in accurate and explainable segmentation. However, the lack of readily available data in echocardiography hampers the training of VLSMs. In this study, we explore using synthetic datasets from Semantic Diffusion Models (SDMs) to enhance VLSMs for echocardiography segmentation. We evaluate results for two popular VLSMs (CLIPSeg and CRIS) using seven different kinds of language prompts derived from several attributes, automatically extracted from echocardiography images, segmentation masks, and their metadata. Our results show improved metrics and faster convergence when pretraining VLSMs on SDM-generated synthetic images before finetuning on real images. The code, configs, and prompts are available at https://github.com/naamiinepal/synthetic-boost.
Exploring Transfer Learning in Medical Image Segmentation using Vision-Language Models
Aug 15, 2023



Abstract:Medical Image Segmentation is crucial in various clinical applications within the medical domain. While state-of-the-art segmentation models have proven effective, integrating textual guidance to enhance visual features for this task remains an area with limited progress. Existing segmentation models that utilize textual guidance are primarily trained on open-domain images, raising concerns about their direct applicability in the medical domain without manual intervention or fine-tuning. To address these challenges, we propose using multimodal vision-language models for capturing semantic information from image descriptions and images, enabling the segmentation of diverse medical images. This study comprehensively evaluates existing vision language models across multiple datasets to assess their transferability from the open domain to the medical field. Furthermore, we introduce variations of image descriptions for previously unseen images in the dataset, revealing notable variations in model performance based on the generated prompts. Our findings highlight the distribution shift between the open-domain images and the medical domain and show that the segmentation models trained on open-domain images are not directly transferrable to the medical field. But their performance can be increased by finetuning them in the medical datasets. We report the zero-shot and finetuned segmentation performance of 4 Vision Language Models (VLMs) on 11 medical datasets using 9 types of prompts derived from 14 attributes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge