Cristian A. Linte
Evaluating Large Vision-language Models for Surgical Tool Detection
Jan 23, 2026Abstract:Surgery is a highly complex process, and artificial intelligence has emerged as a transformative force in supporting surgical guidance and decision-making. However, the unimodal nature of most current AI systems limits their ability to achieve a holistic understanding of surgical workflows. This highlights the need for general-purpose surgical AI systems capable of comprehensively modeling the interrelated components of surgical scenes. Recent advances in large vision-language models that integrate multimodal data processing offer strong potential for modeling surgical tasks and providing human-like scene reasoning and understanding. Despite their promise, systematic investigations of VLMs in surgical applications remain limited. In this study, we evaluate the effectiveness of large VLMs for the fundamental surgical vision task of detecting surgical tools. Specifically, we investigate three state-of-the-art VLMs, Qwen2.5, LLaVA1.5, and InternVL3.5, on the GraSP robotic surgery dataset under both zero-shot and parameter-efficient LoRA fine-tuning settings. Our results demonstrate that Qwen2.5 consistently achieves superior detection performance in both configurations among the evaluated VLMs. Furthermore, compared with the open-set detection baseline Grounding DINO, Qwen2.5 exhibits stronger zero-shot generalization and comparable fine-tuned performance. Notably, Qwen2.5 shows superior instrument recognition, while Grounding DINO demonstrates stronger localization.
Toward Patient-specific Partial Point Cloud to Surface Completion for Pre- to Intra-operative Registration in Image-guided Liver Interventions
May 26, 2025Abstract:Intra-operative data captured during image-guided surgery lacks sub-surface information, where key regions of interest, such as vessels and tumors, reside. Image-to-physical registration enables the fusion of pre-operative information and intra-operative data, typically represented as a point cloud. However, this registration process struggles due to partial visibility of the intra-operative point cloud. In this research, we propose a patient-specific point cloud completion approach to assist with the registration process. Specifically, we leverage VN-OccNet to generate a complete liver surface from a partial intra-operative point cloud. The network is trained in a patient-specific manner, where simulated deformations from the pre-operative model are used to train the model. First, we conduct an in-depth analysis of VN-OccNet's rotation-equivariant property and its effectiveness in recovering complete surfaces from partial intra-operative surfaces. Next, we integrate the completed intra-operative surface into the Go-ICP registration algorithm to demonstrate its utility in improving initial rigid registration outcomes. Our results highlight the promise of this patient-specific completion approach in mitigating the challenges posed by partial intra-operative visibility. The rotation equivariant and surface generation capabilities of VN-OccNet hold strong promise for developing robust registration frameworks for variations of the intra-operative point cloud.
Evaluation of Intra-operative Patient-specific Methods for Point Cloud Completion for Minimally Invasive Liver Interventions
Mar 15, 2025Abstract:The registration between the pre-operative model and the intra-operative surface is crucial in image-guided liver surgery, as it facilitates the effective use of pre-operative information during the procedure. However, the intra-operative surface, usually represented as a point cloud, often has limited coverage, especially in laparoscopic surgery, and is prone to holes and noise, posing significant challenges for registration methods. Point cloud completion methods have the potential to alleviate these issues. Thus, we explore six state-of-the-art point cloud completion methods to identify the optimal completion method for liver surgery applications. We focus on a patient-specific approach for liver point cloud completion from a partial liver surface under three cases: canonical pose, non-canonical pose, and canonical pose with noise. The transformer-based method, AdaPoinTr, outperforms all other methods to generate a complete point cloud from the given partial liver point cloud under the canonical pose. On the other hand, our findings reveal substantial performance degradation of these methods under non-canonical poses and noisy settings, highlighting the limitations of these methods, which suggests the need for a robust point completion method for its application in image-guided liver surgery.
Assessing the Performance of the DINOv2 Self-supervised Learning Vision Transformer Model for the Segmentation of the Left Atrium from MRI Images
Nov 14, 2024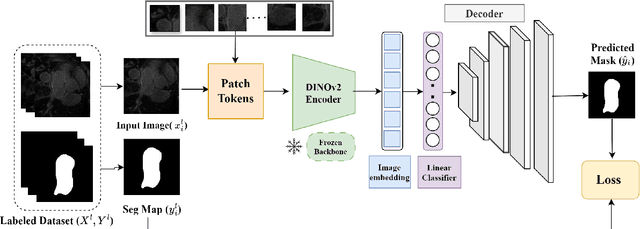
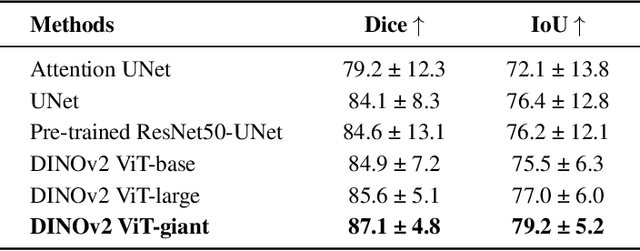
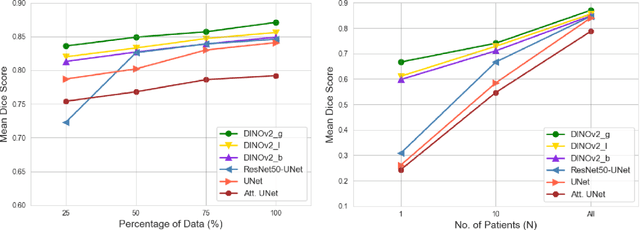
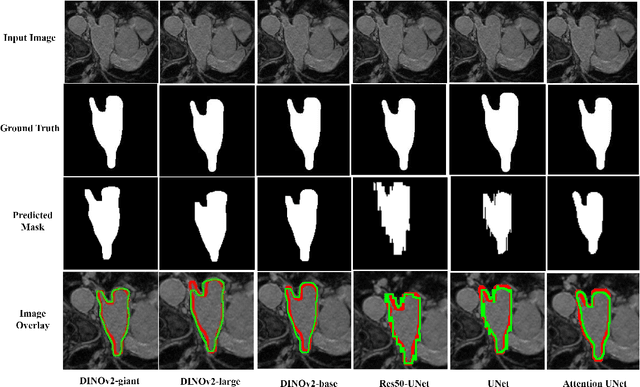
Abstract:Accurate left atrium (LA) segmentation from pre-operative scans is crucial for diagnosing atrial fibrillation, treatment planning, and supporting surgical interventions. While deep learning models are key in medical image segmentation, they often require extensive manually annotated data. Foundation models trained on larger datasets have reduced this dependency, enhancing generalizability and robustness through transfer learning. We explore DINOv2, a self-supervised learning vision transformer trained on natural images, for LA segmentation using MRI. The challenges for LA's complex anatomy, thin boundaries, and limited annotated data make accurate segmentation difficult before & during the image-guided intervention. We demonstrate DINOv2's ability to provide accurate & consistent segmentation, achieving a mean Dice score of .871 & a Jaccard Index of .792 for end-to-end fine-tuning. Through few-shot learning across various data sizes & patient counts, DINOv2 consistently outperforms baseline models. These results suggest that DINOv2 effectively adapts to MRI with limited data, highlighting its potential as a competitive tool for segmentation & encouraging broader use in medical imaging.
Investigating the Robustness of Vision Transformers against Label Noise in Medical Image Classification
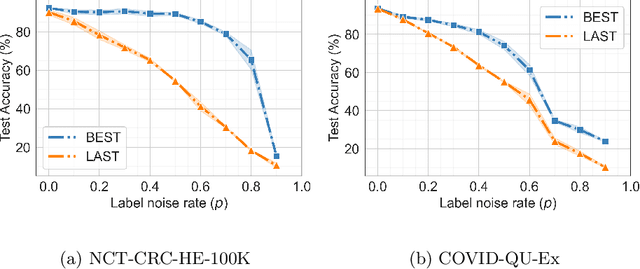
Feb 26, 2024Abstract:Label noise in medical image classification datasets significantly hampers the training of supervised deep learning methods, undermining their generalizability. The test performance of a model tends to decrease as the label noise rate increases. Over recent years, several methods have been proposed to mitigate the impact of label noise in medical image classification and enhance the robustness of the model. Predominantly, these works have employed CNN-based architectures as the backbone of their classifiers for feature extraction. However, in recent years, Vision Transformer (ViT)-based backbones have replaced CNNs, demonstrating improved performance and a greater ability to learn more generalizable features, especially when the dataset is large. Nevertheless, no prior work has rigorously investigated how transformer-based backbones handle the impact of label noise in medical image classification. In this paper, we investigate the architectural robustness of ViT against label noise and compare it to that of CNNs. We use two medical image classification datasets -- COVID-DU-Ex, and NCT-CRC-HE-100K -- both corrupted by injecting label noise at various rates. Additionally, we show that pretraining is crucial for ensuring ViT's improved robustness against label noise in supervised training.
Medical Vision Language Pretraining: A survey
Dec 11, 2023Abstract:Medical Vision Language Pretraining (VLP) has recently emerged as a promising solution to the scarcity of labeled data in the medical domain. By leveraging paired/unpaired vision and text datasets through self-supervised learning, models can be trained to acquire vast knowledge and learn robust feature representations. Such pretrained models have the potential to enhance multiple downstream medical tasks simultaneously, reducing the dependency on labeled data. However, despite recent progress and its potential, there is no such comprehensive survey paper that has explored the various aspects and advancements in medical VLP. In this paper, we specifically review existing works through the lens of different pretraining objectives, architectures, downstream evaluation tasks, and datasets utilized for pretraining and downstream tasks. Subsequently, we delve into current challenges in medical VLP, discussing existing and potential solutions, and conclude by highlighting future directions. To the best of our knowledge, this is the first survey focused on medical VLP.
Improving Medical Image Classification in Noisy Labels Using Only Self-supervised Pretraining
Aug 08, 2023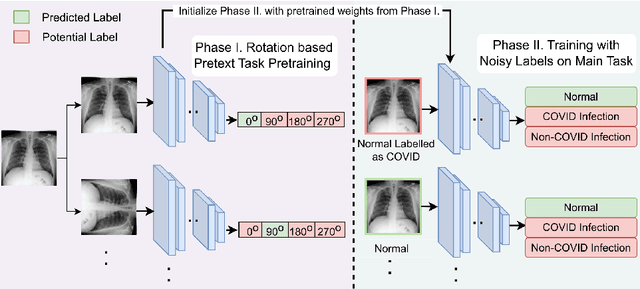

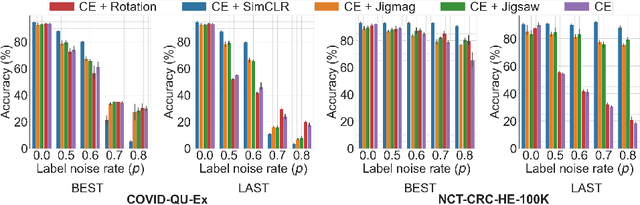
Abstract:Noisy labels hurt deep learning-based supervised image classification performance as the models may overfit the noise and learn corrupted feature extractors. For natural image classification training with noisy labeled data, model initialization with contrastive self-supervised pretrained weights has shown to reduce feature corruption and improve classification performance. However, no works have explored: i) how other self-supervised approaches, such as pretext task-based pretraining, impact the learning with noisy label, and ii) any self-supervised pretraining methods alone for medical images in noisy label settings. Medical images often feature smaller datasets and subtle inter class variations, requiring human expertise to ensure correct classification. Thus, it is not clear if the methods improving learning with noisy labels in natural image datasets such as CIFAR would also help with medical images. In this work, we explore contrastive and pretext task-based self-supervised pretraining to initialize the weights of a deep learning classification model for two medical datasets with self-induced noisy labels -- NCT-CRC-HE-100K tissue histological images and COVID-QU-Ex chest X-ray images. Our results show that models initialized with pretrained weights obtained from self-supervised learning can effectively learn better features and improve robustness against noisy labels.
M-VAAL: Multimodal Variational Adversarial Active Learning for Downstream Medical Image Analysis Tasks
Jun 21, 2023Abstract:Acquiring properly annotated data is expensive in the medical field as it requires experts, time-consuming protocols, and rigorous validation. Active learning attempts to minimize the need for large annotated samples by actively sampling the most informative examples for annotation. These examples contribute significantly to improving the performance of supervised machine learning models, and thus, active learning can play an essential role in selecting the most appropriate information in deep learning-based diagnosis, clinical assessments, and treatment planning. Although some existing works have proposed methods for sampling the best examples for annotation in medical image analysis, they are not task-agnostic and do not use multimodal auxiliary information in the sampler, which has the potential to increase robustness. Therefore, in this work, we propose a Multimodal Variational Adversarial Active Learning (M-VAAL) method that uses auxiliary information from additional modalities to enhance the active sampling. We applied our method to two datasets: i) brain tumor segmentation and multi-label classification using the BraTS2018 dataset, and ii) chest X-ray image classification using the COVID-QU-Ex dataset. Our results show a promising direction toward data-efficient learning under limited annotations.
A Disparity Refinement Framework for Learning-based Stereo Matching Methods in Cross-domain Setting for Laparoscopic Images
Feb 05, 2023Abstract:Purpose: Stereo matching methods that enable depth estimation are crucial for visualization enhancement applications in computer-assisted surgery (CAS). Learning-based stereo matching methods are promising to predict accurate results on laparoscopic images. However, they require a large amount of training data, and their performance may be degraded due to domain shifts. Methods: Maintaining robustness and improving the accuracy of learning-based methods are still open problems. To overcome the limitations of learning-based methods, we propose a disparity refinement framework consisting of a local disparity refinement method and a global disparity refinement method to improve the results of learning-based stereo matching methods in a cross-domain setting. Those learning-based stereo matching methods are pre-trained on a large public dataset of natural images and are tested on two datasets of laparoscopic images. Results: Qualitative and quantitative results suggest that our proposed disparity framework can effectively refine disparity maps when they are noise-corrupted on an unseen dataset, without compromising prediction accuracy when the network can generalize well on an unseen dataset. Conclusion: Our proposed disparity refinement framework could work with learning-based methods to achieve robust and accurate disparity prediction. Yet, as a large laparoscopic dataset for training learning-based methods does not exist and the generalization ability of networks remains to be improved, the incorporation of the proposed disparity refinement framework into existing networks will contribute to improving their overall accuracy and robustness associated with depth estimation.
Learning Feature Descriptors for Pre- and Intra-operative Point Cloud Matching for Laparoscopic Liver Registration
Nov 07, 2022Abstract:Purpose: In laparoscopic liver surgery (LLS), pre-operative information can be overlaid onto the intra-operative scene by registering a 3D pre-operative model to the intra-operative partial surface reconstructed from the laparoscopic video. To assist with this task, we explore the use of learning-based feature descriptors, which, to our best knowledge, have not been explored for use in laparoscopic liver registration. Furthermore, a dataset to train and evaluate the use of learning-based descriptors does not exist. Methods: We present the LiverMatch dataset consisting of 16 preoperative models and their simulated intra-operative 3D surfaces. We also propose the LiverMatch network designed for this task, which outputs per-point feature descriptors, visibility scores, and matched points. Results: We compare the proposed LiverMatch network with anetwork closest to LiverMatch, and a histogram-based 3D descriptor on the testing split of the LiverMatch dataset, which includes two unseen pre-operative models and 1400 intra-operative surfaces. Results suggest that our LiverMatch network can predict more accurate and dense matches than the other two methods and can be seamlessly integrated with a RANSAC-ICP-based registration algorithm to achieve an accurate initial alignment. Conclusion: The use of learning-based feature descriptors in LLR is promising, as it can help achieve an accurate initial rigid alignment, which, in turn, serves as an initialization for subsequent non-rigid registration. We will release the dataset and code upon acceptance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge