Sanjay Bhandari
Hallucination-Aware Multimodal Benchmark for Gastrointestinal Image Analysis with Large Vision-Language Models
May 11, 2025Abstract:Vision-Language Models (VLMs) are becoming increasingly popular in the medical domain, bridging the gap between medical images and clinical language. Existing VLMs demonstrate an impressive ability to comprehend medical images and text queries to generate detailed, descriptive diagnostic medical reports. However, hallucination--the tendency to generate descriptions that are inconsistent with the visual content--remains a significant issue in VLMs, with particularly severe implications in the medical field. To facilitate VLM research on gastrointestinal (GI) image analysis and study hallucination, we curate a multimodal image-text GI dataset: Gut-VLM. This dataset is created using a two-stage pipeline: first, descriptive medical reports of Kvasir-v2 images are generated using ChatGPT, which introduces some hallucinated or incorrect texts. In the second stage, medical experts systematically review these reports, and identify and correct potential inaccuracies to ensure high-quality, clinically reliable annotations. Unlike traditional datasets that contain only descriptive texts, our dataset also features tags identifying hallucinated sentences and their corresponding corrections. A common approach to reducing hallucination in VLM is to finetune the model on a small-scale, problem-specific dataset. However, we take a different strategy using our dataset. Instead of finetuning the VLM solely for generating textual reports, we finetune it to detect and correct hallucinations, an approach we call hallucination-aware finetuning. Our results show that this approach is better than simply finetuning for descriptive report generation. Additionally, we conduct an extensive evaluation of state-of-the-art VLMs across several metrics, establishing a benchmark. GitHub Repo: https://github.com/bhattarailab/Hallucination-Aware-VLM.
NCDD: Nearest Centroid Distance Deficit for Out-Of-Distribution Detection in Gastrointestinal Vision
Dec 02, 2024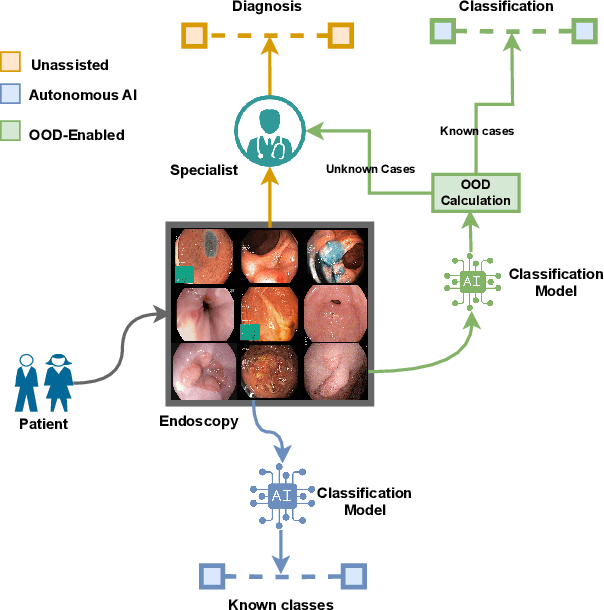

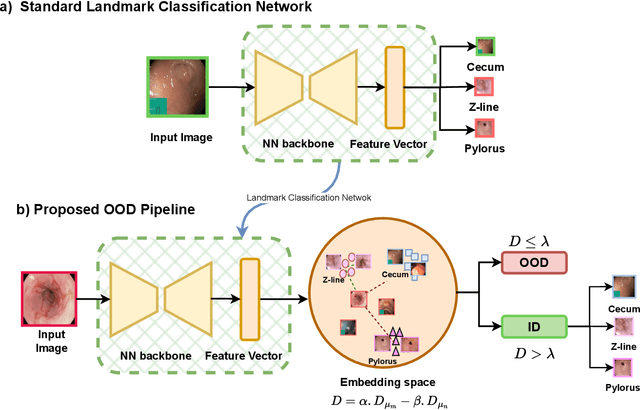
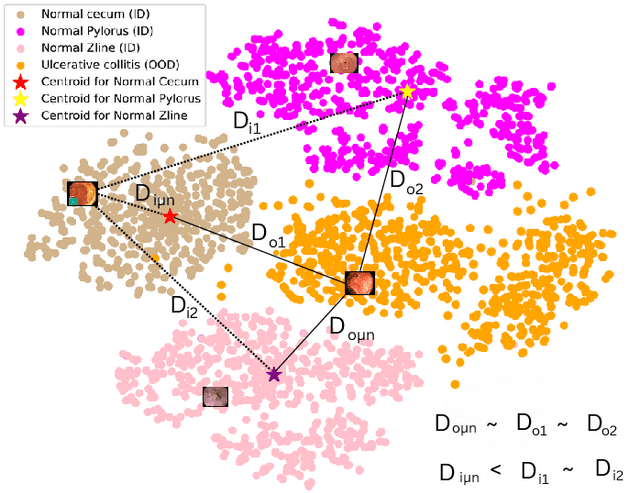
Abstract:The integration of deep learning tools in gastrointestinal vision holds the potential for significant advancements in diagnosis, treatment, and overall patient care. A major challenge, however, is these tools' tendency to make overconfident predictions, even when encountering unseen or newly emerging disease patterns, undermining their reliability. We address this critical issue of reliability by framing it as an out-of-distribution (OOD) detection problem, where previously unseen and emerging diseases are identified as OOD examples. However, gastrointestinal images pose a unique challenge due to the overlapping feature representations between in- Distribution (ID) and OOD examples. Existing approaches often overlook this characteristic, as they are primarily developed for natural image datasets, where feature distinctions are more apparent. Despite the overlap, we hypothesize that the features of an in-distribution example will cluster closer to the centroids of their ground truth class, resulting in a shorter distance to the nearest centroid. In contrast, OOD examples maintain an equal distance from all class centroids. Based on this observation, we propose a novel nearest-centroid distance deficit (NCCD) score in the feature space for gastrointestinal OOD detection. Evaluations across multiple deep learning architectures and two publicly available benchmarks, Kvasir2 and Gastrovision, demonstrate the effectiveness of our approach compared to several state-of-the-art methods. The code and implementation details are publicly available at: https://github.com/bhattarailab/NCDD
TTA-OOD: Test-time Augmentation for Improving Out-of-Distribution Detection in Gastrointestinal Vision
Jul 19, 2024



Abstract:Deep learning has significantly advanced the field of gastrointestinal vision, enhancing disease diagnosis capabilities. One major challenge in automating diagnosis within gastrointestinal settings is the detection of abnormal cases in endoscopic images. Due to the sparsity of data, this process of distinguishing normal from abnormal cases has faced significant challenges, particularly with rare and unseen conditions. To address this issue, we frame abnormality detection as an out-of-distribution (OOD) detection problem. In this setup, a model trained on In-Distribution (ID) data, which represents a healthy GI tract, can accurately identify healthy cases, while abnormalities are detected as OOD, regardless of their class. We introduce a test-time augmentation segment into the OOD detection pipeline, which enhances the distinction between ID and OOD examples, thereby improving the effectiveness of existing OOD methods with the same model. This augmentation shifts the pixel space, which translates into a more distinct semantic representation for OOD examples compared to ID examples. We evaluated our method against existing state-of-the-art OOD scores, showing improvements with test-time augmentation over the baseline approach.
Data Augmentation through Pseudolabels in Automatic Region Based Coronary Artery Segmentation for Disease Diagnosis
Oct 08, 2023Abstract:Coronary Artery Diseases(CADs) though preventable are one of the leading causes of death and disability. Diagnosis of these diseases is often difficult and resource intensive. Segmentation of arteries in angiographic images has evolved as a tool for assistance, helping clinicians in making accurate diagnosis. However, due to the limited amount of data and the difficulty in curating a dataset, the task of segmentation has proven challenging. In this study, we introduce the idea of using pseudolabels as a data augmentation technique to improve the performance of the baseline Yolo model. This method increases the F1 score of the baseline by 9% in the validation dataset and by 3% in the test dataset.
ConvNeXtv2 Fusion with Mask R-CNN for Automatic Region Based Coronary Artery Stenosis Detection for Disease Diagnosis
Oct 07, 2023



Abstract:Coronary Artery Diseases although preventable are one of the leading cause of mortality worldwide. Due to the onerous nature of diagnosis, tackling CADs has proved challenging. This study addresses the automation of resource-intensive and time-consuming process of manually detecting stenotic lesions in coronary arteries in X-ray coronary angiography images. To overcome this challenge, we employ a specialized Convnext-V2 backbone based Mask RCNN model pre-trained for instance segmentation tasks. Our empirical findings affirm that the proposed model exhibits commendable performance in identifying stenotic lesions. Notably, our approach achieves a substantial F1 score of 0.5353 in this demanding task, underscoring its effectiveness in streamlining this intensive process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge