Matthew J. Clarkson
EndoSfM3D: Learning to 3D Reconstruct Any Endoscopic Surgery Scene using Self-supervised Foundation Model
Oct 25, 2025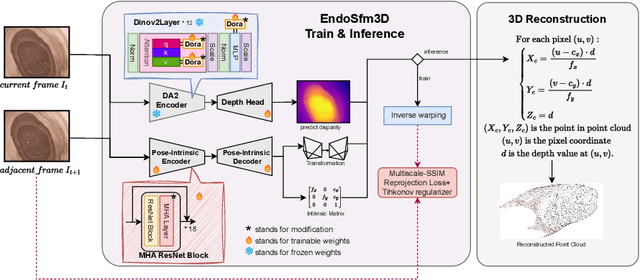
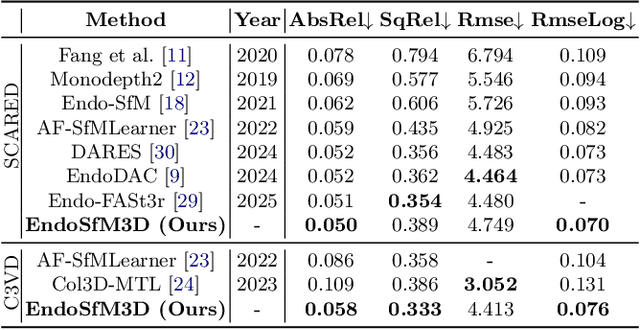
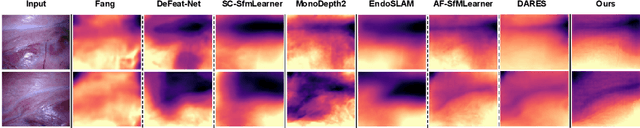

Abstract:3D reconstruction of endoscopic surgery scenes plays a vital role in enhancing scene perception, enabling AR visualization, and supporting context-aware decision-making in image-guided surgery. A critical yet challenging step in this process is the accurate estimation of the endoscope's intrinsic parameters. In real surgical settings, intrinsic calibration is hindered by sterility constraints and the use of specialized endoscopes with continuous zoom and telescope rotation. Most existing methods for endoscopic 3D reconstruction do not estimate intrinsic parameters, limiting their effectiveness for accurate and reliable reconstruction. In this paper, we integrate intrinsic parameter estimation into a self-supervised monocular depth estimation framework by adapting the Depth Anything V2 (DA2) model for joint depth, pose, and intrinsics prediction. We introduce an attention-based pose network and a Weight-Decomposed Low-Rank Adaptation (DoRA) strategy for efficient fine-tuning of DA2. Our method is validated on the SCARED and C3VD public datasets, demonstrating superior performance compared to recent state-of-the-art approaches in self-supervised monocular depth estimation and 3D reconstruction. Code and model weights can be found in project repository: https://github.com/MOYF-beta/EndoSfM3D.
SurgicalVLM-Agent: Towards an Interactive AI Co-Pilot for Pituitary Surgery
Mar 12, 2025Abstract:Image-guided surgery demands adaptive, real-time decision support, yet static AI models struggle with structured task planning and providing interactive guidance. Large vision-language models (VLMs) offer a promising solution by enabling dynamic task planning and predictive decision support. We introduce SurgicalVLM-Agent, an AI co-pilot for image-guided pituitary surgery, capable of conversation, planning, and task execution. The agent dynamically processes surgeon queries and plans the tasks such as MRI tumor segmentation, endoscope anatomy segmentation, overlaying preoperative imaging with intraoperative views, instrument tracking, and surgical visual question answering (VQA). To enable structured task planning, we develop the PitAgent dataset, a surgical context-aware dataset covering segmentation, overlaying, instrument localization, tool tracking, tool-tissue interactions, phase identification, and surgical activity recognition. Additionally, we propose FFT-GaLore, a fast Fourier transform (FFT)-based gradient projection technique for efficient low-rank adaptation, optimizing fine-tuning for LLaMA 3.2 in surgical environments. We validate SurgicalVLM-Agent by assessing task planning and prompt generation on our PitAgent dataset and evaluating zero-shot VQA using a public pituitary dataset. Results demonstrate state-of-the-art performance in task planning and query interpretation, with highly semantically meaningful VQA responses, advancing AI-driven surgical assistance.
PitVQA++: Vector Matrix-Low-Rank Adaptation for Open-Ended Visual Question Answering in Pituitary Surgery
Feb 19, 2025Abstract:Vision-Language Models (VLMs) in visual question answering (VQA) offer a unique opportunity to enhance intra-operative decision-making, promote intuitive interactions, and significantly advancing surgical education. However, the development of VLMs for surgical VQA is challenging due to limited datasets and the risk of overfitting and catastrophic forgetting during full fine-tuning of pretrained weights. While parameter-efficient techniques like Low-Rank Adaptation (LoRA) and Matrix of Rank Adaptation (MoRA) address adaptation challenges, their uniform parameter distribution overlooks the feature hierarchy in deep networks, where earlier layers, that learn general features, require more parameters than later ones. This work introduces PitVQA++ with an open-ended PitVQA dataset and vector matrix-low-rank adaptation (Vector-MoLoRA), an innovative VLM fine-tuning approach for adapting GPT-2 to pituitary surgery. Open-Ended PitVQA comprises around 101,803 frames from 25 procedural videos with 745,972 question-answer sentence pairs, covering key surgical elements such as phase and step recognition, context understanding, tool detection, localization, and interactions recognition. Vector-MoLoRA incorporates the principles of LoRA and MoRA to develop a matrix-low-rank adaptation strategy that employs vector ranking to allocate more parameters to earlier layers, gradually reducing them in the later layers. Our approach, validated on the Open-Ended PitVQA and EndoVis18-VQA datasets, effectively mitigates catastrophic forgetting while significantly enhancing performance over recent baselines. Furthermore, our risk-coverage analysis highlights its enhanced reliability and trustworthiness in handling uncertain predictions. Our source code and dataset is available at~\url{https://github.com/HRL-Mike/PitVQA-Plus}.
Automated Surgical Skill Assessment in Endoscopic Pituitary Surgery using Real-time Instrument Tracking on a High-fidelity Bench-top Phantom
Sep 25, 2024Abstract:Improved surgical skill is generally associated with improved patient outcomes, although assessment is subjective; labour-intensive; and requires domain specific expertise. Automated data driven metrics can alleviate these difficulties, as demonstrated by existing machine learning instrument tracking models in minimally invasive surgery. However, these models have been tested on limited datasets of laparoscopic surgery, with a focus on isolated tasks and robotic surgery. In this paper, a new public dataset is introduced, focusing on simulated surgery, using the nasal phase of endoscopic pituitary surgery as an exemplar. Simulated surgery allows for a realistic yet repeatable environment, meaning the insights gained from automated assessment can be used by novice surgeons to hone their skills on the simulator before moving to real surgery. PRINTNet (Pituitary Real-time INstrument Tracking Network) has been created as a baseline model for this automated assessment. Consisting of DeepLabV3 for classification and segmentation; StrongSORT for tracking; and the NVIDIA Holoscan SDK for real-time performance, PRINTNet achieved 71.9% Multiple Object Tracking Precision running at 22 Frames Per Second. Using this tracking output, a Multilayer Perceptron achieved 87% accuracy in predicting surgical skill level (novice or expert), with the "ratio of total procedure time to instrument visible time" correlated with higher surgical skill. This therefore demonstrates the feasibility of automated surgical skill assessment in simulated endoscopic pituitary surgery. The new publicly available dataset can be found here: https://doi.org/10.5522/04/26511049.
DARES: Depth Anything in Robotic Endoscopic Surgery with Self-supervised Vector-LoRA of the Foundation Model
Aug 30, 2024



Abstract:Robotic-assisted surgery (RAS) relies on accurate depth estimation for 3D reconstruction and visualization. While foundation models like Depth Anything Models (DAM) show promise, directly applying them to surgery often yields suboptimal results. Fully fine-tuning on limited surgical data can cause overfitting and catastrophic forgetting, compromising model robustness and generalization. Although Low-Rank Adaptation (LoRA) addresses some adaptation issues, its uniform parameter distribution neglects the inherent feature hierarchy, where earlier layers, learning more general features, require more parameters than later ones. To tackle this issue, we introduce Depth Anything in Robotic Endoscopic Surgery (DARES), a novel approach that employs a new adaptation technique, Vector Low-Rank Adaptation (Vector-LoRA) on the DAM V2 to perform self-supervised monocular depth estimation in RAS scenes. To enhance learning efficiency, we introduce Vector-LoRA by integrating more parameters in earlier layers and gradually decreasing parameters in later layers. We also design a reprojection loss based on the multi-scale SSIM error to enhance depth perception by better tailoring the foundation model to the specific requirements of the surgical environment. The proposed method is validated on the SCARED dataset and demonstrates superior performance over recent state-of-the-art self-supervised monocular depth estimation techniques, achieving an improvement of 13.3% in the absolute relative error metric. The code and pre-trained weights are available at https://github.com/mobarakol/DARES.
HUP-3D: A 3D multi-view synthetic dataset for assisted-egocentric hand-ultrasound pose estimation
Jul 12, 2024Abstract:We present HUP-3D, a 3D multi-view multi-modal synthetic dataset for hand-ultrasound (US) probe pose estimation in the context of obstetric ultrasound. Egocentric markerless 3D joint pose estimation has potential applications in mixed reality based medical education. The ability to understand hand and probe movements programmatically opens the door to tailored guidance and mentoring applications. Our dataset consists of over 31k sets of RGB, depth and segmentation mask frames, including pose related ground truth data, with a strong emphasis on image diversity and complexity. Adopting a camera viewpoint-based sphere concept allows us to capture a variety of views and generate multiple hand grasp poses using a pre-trained network. Additionally, our approach includes a software-based image rendering concept, enhancing diversity with various hand and arm textures, lighting conditions, and background images. Furthermore, we validated our proposed dataset with state-of-the-art learning models and we obtained the lowest hand-object keypoint errors. The dataset and other details are provided with the supplementary material. The source code of our grasp generation and rendering pipeline will be made publicly available.
Nonrigid Reconstruction of Freehand Ultrasound without a Tracker
Jul 08, 2024Abstract:Reconstructing 2D freehand Ultrasound (US) frames into 3D space without using a tracker has recently seen advances with deep learning. Predicting good frame-to-frame rigid transformations is often accepted as the learning objective, especially when the ground-truth labels from spatial tracking devices are inherently rigid transformations. Motivated by a) the observed nonrigid deformation due to soft tissue motion during scanning, and b) the highly sensitive prediction of rigid transformation, this study investigates the methods and their benefits in predicting nonrigid transformations for reconstructing 3D US. We propose a novel co-optimisation algorithm for simultaneously estimating rigid transformations among US frames, supervised by ground-truth from a tracker, and a nonrigid deformation, optimised by a regularised registration network. We show that these two objectives can be either optimised using meta-learning or combined by weighting. A fast scattered data interpolation is also developed for enabling frequent reconstruction and registration of non-parallel US frames, during training. With a new data set containing over 357,000 frames in 720 scans, acquired from 60 subjects, the experiments demonstrate that, due to an expanded thus easier-to-optimise solution space, the generalisation is improved with the added deformation estimation, with respect to the rigid ground-truth. The global pixel reconstruction error (assessing accumulative prediction) is lowered from 18.48 to 16.51 mm, compared with baseline rigid-transformation-predicting methods. Using manually identified landmarks, the proposed co-optimisation also shows potentials in compensating nonrigid tissue motion at inference, which is not measurable by tracker-provided ground-truth. The code and data used in this paper are made publicly available at https://github.com/QiLi111/NR-Rec-FUS.
Competing for pixels: a self-play algorithm for weakly-supervised segmentation
May 26, 2024Abstract:Weakly-supervised segmentation (WSS) methods, reliant on image-level labels indicating object presence, lack explicit correspondence between labels and regions of interest (ROIs), posing a significant challenge. Despite this, WSS methods have attracted attention due to their much lower annotation costs compared to fully-supervised segmentation. Leveraging reinforcement learning (RL) self-play, we propose a novel WSS method that gamifies image segmentation of a ROI. We formulate segmentation as a competition between two agents that compete to select ROI-containing patches until exhaustion of all such patches. The score at each time-step, used to compute the reward for agent training, represents likelihood of object presence within the selection, determined by an object presence detector pre-trained using only image-level binary classification labels of object presence. Additionally, we propose a game termination condition that can be called by either side upon exhaustion of all ROI-containing patches, followed by the selection of a final patch from each. Upon termination, the agent is incentivised if ROI-containing patches are exhausted or disincentivised if an ROI-containing patch is found by the competitor. This competitive setup ensures minimisation of over- or under-segmentation, a common problem with WSS methods. Extensive experimentation across four datasets demonstrates significant performance improvements over recent state-of-the-art methods. Code: https://github.com/s-sd/spurl/tree/main/wss
PitVQA: Image-grounded Text Embedding LLM for Visual Question Answering in Pituitary Surgery
May 22, 2024



Abstract:Visual Question Answering (VQA) within the surgical domain, utilizing Large Language Models (LLMs), offers a distinct opportunity to improve intra-operative decision-making and facilitate intuitive surgeon-AI interaction. However, the development of LLMs for surgical VQA is hindered by the scarcity of diverse and extensive datasets with complex reasoning tasks. Moreover, contextual fusion of the image and text modalities remains an open research challenge due to the inherent differences between these two types of information and the complexity involved in aligning them. This paper introduces PitVQA, a novel dataset specifically designed for VQA in endonasal pituitary surgery and PitVQA-Net, an adaptation of the GPT2 with a novel image-grounded text embedding for surgical VQA. PitVQA comprises 25 procedural videos and a rich collection of question-answer pairs spanning crucial surgical aspects such as phase and step recognition, context understanding, tool detection and localization, and tool-tissue interactions. PitVQA-Net consists of a novel image-grounded text embedding that projects image and text features into a shared embedding space and GPT2 Backbone with an excitation block classification head to generate contextually relevant answers within the complex domain of endonasal pituitary surgery. Our image-grounded text embedding leverages joint embedding, cross-attention and contextual representation to understand the contextual relationship between questions and surgical images. We demonstrate the effectiveness of PitVQA-Net on both the PitVQA and the publicly available EndoVis18-VQA dataset, achieving improvements in balanced accuracy of 8% and 9% over the most recent baselines, respectively. Our code and dataset is available at https://github.com/mobarakol/PitVQA.
Surgical-DeSAM: Decoupling SAM for Instrument Segmentation in Robotic Surgery
Apr 22, 2024Abstract:Purpose: The recent Segment Anything Model (SAM) has demonstrated impressive performance with point, text or bounding box prompts, in various applications. However, in safety-critical surgical tasks, prompting is not possible due to (i) the lack of per-frame prompts for supervised learning, (ii) it is unrealistic to prompt frame-by-frame in a real-time tracking application, and (iii) it is expensive to annotate prompts for offline applications. Methods: We develop Surgical-DeSAM to generate automatic bounding box prompts for decoupling SAM to obtain instrument segmentation in real-time robotic surgery. We utilise a commonly used detection architecture, DETR, and fine-tuned it to obtain bounding box prompt for the instruments. We then empolyed decoupling SAM (DeSAM) by replacing the image encoder with DETR encoder and fine-tune prompt encoder and mask decoder to obtain instance segmentation for the surgical instruments. To improve detection performance, we adopted the Swin-transformer to better feature representation. Results: The proposed method has been validated on two publicly available datasets from the MICCAI surgical instruments segmentation challenge EndoVis 2017 and 2018. The performance of our method is also compared with SOTA instrument segmentation methods and demonstrated significant improvements with dice metrics of 89.62 and 90.70 for the EndoVis 2017 and 2018. Conclusion: Our extensive experiments and validations demonstrate that Surgical-DeSAM enables real-time instrument segmentation without any additional prompting and outperforms other SOTA segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge