Ziyi Shen
SAMReg: SAM-enabled Image Registration with ROI-based Correspondence
Oct 17, 2024



Abstract:This paper describes a new spatial correspondence representation based on paired regions-of-interest (ROIs), for medical image registration. The distinct properties of the proposed ROI-based correspondence are discussed, in the context of potential benefits in clinical applications following image registration, compared with alternative correspondence-representing approaches, such as those based on sampled displacements and spatial transformation functions. These benefits include a clear connection between learning-based image registration and segmentation, which in turn motivates two cases of image registration approaches using (pre-)trained segmentation networks. Based on the segment anything model (SAM), a vision foundation model for segmentation, we develop a new registration algorithm SAMReg, which does not require any training (or training data), gradient-based fine-tuning or prompt engineering. The proposed SAMReg models are evaluated across five real-world applications, including intra-subject registration tasks with cardiac MR and lung CT, challenging inter-subject registration scenarios with prostate MR and retinal imaging, and an additional evaluation with a non-clinical example with aerial image registration. The proposed methods outperform both intensity-based iterative algorithms and DDF-predicting learning-based networks across tested metrics including Dice and target registration errors on anatomical structures, and further demonstrates competitive performance compared to weakly-supervised registration approaches that rely on fully-segmented training data. Open source code and examples are available at: https://github.com/sqhuang0103/SAMReg.git.
Nonrigid Reconstruction of Freehand Ultrasound without a Tracker
Jul 08, 2024Abstract:Reconstructing 2D freehand Ultrasound (US) frames into 3D space without using a tracker has recently seen advances with deep learning. Predicting good frame-to-frame rigid transformations is often accepted as the learning objective, especially when the ground-truth labels from spatial tracking devices are inherently rigid transformations. Motivated by a) the observed nonrigid deformation due to soft tissue motion during scanning, and b) the highly sensitive prediction of rigid transformation, this study investigates the methods and their benefits in predicting nonrigid transformations for reconstructing 3D US. We propose a novel co-optimisation algorithm for simultaneously estimating rigid transformations among US frames, supervised by ground-truth from a tracker, and a nonrigid deformation, optimised by a regularised registration network. We show that these two objectives can be either optimised using meta-learning or combined by weighting. A fast scattered data interpolation is also developed for enabling frequent reconstruction and registration of non-parallel US frames, during training. With a new data set containing over 357,000 frames in 720 scans, acquired from 60 subjects, the experiments demonstrate that, due to an expanded thus easier-to-optimise solution space, the generalisation is improved with the added deformation estimation, with respect to the rigid ground-truth. The global pixel reconstruction error (assessing accumulative prediction) is lowered from 18.48 to 16.51 mm, compared with baseline rigid-transformation-predicting methods. Using manually identified landmarks, the proposed co-optimisation also shows potentials in compensating nonrigid tissue motion at inference, which is not measurable by tracker-provided ground-truth. The code and data used in this paper are made publicly available at https://github.com/QiLi111/NR-Rec-FUS.
Poisson Ordinal Network for Gleason Group Estimation Using Bi-Parametric MRI
Jul 08, 2024



Abstract:The Gleason groups serve as the primary histological grading system for prostate cancer, providing crucial insights into the cancer's potential for growth and metastasis. In clinical practice, pathologists determine the Gleason groups based on specimens obtained from ultrasound-guided biopsies. In this study, we investigate the feasibility of directly estimating the Gleason groups from MRI scans to reduce otherwise required biopsies. We identify two characteristics of this task, ordinality and the resulting dependent yet unknown variances between Gleason groups. In addition to the inter- / intra- observer variability in a multi-step Gleason scoring process based on the interpretation of Gleason patterns, our MR-based prediction is also subject to specimen sampling variance and, to a lesser degree, varying MR imaging protocols. To address this challenge, we propose a novel Poisson ordinal network (PON). PONs model the prediction using a Poisson distribution and leverages Poisson encoding and Poisson focal loss to capture a learnable dependency between ordinal classes (here, Gleason groups), rather than relying solely on the numerical ground-truth (e.g. Gleason Groups 1-5 or Gleason Scores 6-10). To improve this modelling efficacy, PONs also employ contrastive learning with a memory bank to regularise intra-class variance, decoupling the memory requirement of contrast learning from the batch size. Experimental results based on the images labelled by saturation biopsies from 265 prior-biopsy-blind patients, across two tasks demonstrate the superiority and effectiveness of our proposed method.
One registration is worth two segmentations
May 17, 2024



Abstract:The goal of image registration is to establish spatial correspondence between two or more images, traditionally through dense displacement fields (DDFs) or parametric transformations (e.g., rigid, affine, and splines). Rethinking the existing paradigms of achieving alignment via spatial transformations, we uncover an alternative but more intuitive correspondence representation: a set of corresponding regions-of-interest (ROI) pairs, which we demonstrate to have sufficient representational capability as other correspondence representation methods.Further, it is neither necessary nor sufficient for these ROIs to hold specific anatomical or semantic significance. In turn, we formulate image registration as searching for the same set of corresponding ROIs from both moving and fixed images - in other words, two multi-class segmentation tasks on a pair of images. For a general-purpose and practical implementation, we integrate the segment anything model (SAM) into our proposed algorithms, resulting in a SAM-enabled registration (SAMReg) that does not require any training data, gradient-based fine-tuning or engineered prompts. We experimentally show that the proposed SAMReg is capable of segmenting and matching multiple ROI pairs, which establish sufficiently accurate correspondences, in three clinical applications of registering prostate MR, cardiac MR and abdominal CT images. Based on metrics including Dice and target registration errors on anatomical structures, the proposed registration outperforms both intensity-based iterative algorithms and DDF-predicting learning-based networks, even yielding competitive performance with weakly-supervised registration which requires fully-segmented training data.
Long-term Dependency for 3D Reconstruction of Freehand Ultrasound Without External Tracker
Oct 16, 2023Abstract:Objective: Reconstructing freehand ultrasound in 3D without any external tracker has been a long-standing challenge in ultrasound-assisted procedures. We aim to define new ways of parameterising long-term dependencies, and evaluate the performance. Methods: First, long-term dependency is encoded by transformation positions within a frame sequence. This is achieved by combining a sequence model with a multi-transformation prediction. Second, two dependency factors are proposed, anatomical image content and scanning protocol, for contributing towards accurate reconstruction. Each factor is quantified experimentally by reducing respective training variances. Results: 1) The added long-term dependency up to 400 frames at 20 frames per second (fps) indeed improved reconstruction, with an up to 82.4% lowered accumulated error, compared with the baseline performance. The improvement was found to be dependent on sequence length, transformation interval and scanning protocol and, unexpectedly, not on the use of recurrent networks with long-short term modules; 2) Decreasing either anatomical or protocol variance in training led to poorer reconstruction accuracy. Interestingly, greater performance was gained from representative protocol patterns, than from representative anatomical features. Conclusion: The proposed algorithm uses hyperparameter tuning to effectively utilise long-term dependency. The proposed dependency factors are of practical significance in collecting diverse training data, regulating scanning protocols and developing efficient networks. Significance: The proposed new methodology with publicly available volunteer data and code for parametersing the long-term dependency, experimentally shown to be valid sources of performance improvement, which could potentially lead to better model development and practical optimisation of the reconstruction application.
Privileged Anatomical and Protocol Discrimination in Trackerless 3D Ultrasound Reconstruction
Aug 20, 2023Abstract:Three-dimensional (3D) freehand ultrasound (US) reconstruction without using any additional external tracking device has seen recent advances with deep neural networks (DNNs). In this paper, we first investigated two identified contributing factors of the learned inter-frame correlation that enable the DNN-based reconstruction: anatomy and protocol. We propose to incorporate the ability to represent these two factors - readily available during training - as the privileged information to improve existing DNN-based methods. This is implemented in a new multi-task method, where the anatomical and protocol discrimination are used as auxiliary tasks. We further develop a differentiable network architecture to optimise the branching location of these auxiliary tasks, which controls the ratio between shared and task-specific network parameters, for maximising the benefits from the two auxiliary tasks. Experimental results, on a dataset with 38 forearms of 19 volunteers acquired with 6 different scanning protocols, show that 1) both anatomical and protocol variances are enabling factors for DNN-based US reconstruction; 2) learning how to discriminate different subjects (anatomical variance) and predefined types of scanning paths (protocol variance) both significantly improve frame prediction accuracy, volume reconstruction overlap, accumulated tracking error and final drift, using the proposed algorithm.
Combiner and HyperCombiner Networks: Rules to Combine Multimodality MR Images for Prostate Cancer Localisation
Jul 17, 2023



Abstract:One of the distinct characteristics in radiologists' reading of multiparametric prostate MR scans, using reporting systems such as PI-RADS v2.1, is to score individual types of MR modalities, T2-weighted, diffusion-weighted, and dynamic contrast-enhanced, and then combine these image-modality-specific scores using standardised decision rules to predict the likelihood of clinically significant cancer. This work aims to demonstrate that it is feasible for low-dimensional parametric models to model such decision rules in the proposed Combiner networks, without compromising the accuracy of predicting radiologic labels: First, it is shown that either a linear mixture model or a nonlinear stacking model is sufficient to model PI-RADS decision rules for localising prostate cancer. Second, parameters of these (generalised) linear models are proposed as hyperparameters, to weigh multiple networks that independently represent individual image modalities in the Combiner network training, as opposed to end-to-end modality ensemble. A HyperCombiner network is developed to train a single image segmentation network that can be conditioned on these hyperparameters during inference, for much improved efficiency. Experimental results based on data from 850 patients, for the application of automating radiologist labelling multi-parametric MR, compare the proposed combiner networks with other commonly-adopted end-to-end networks. Using the added advantages of obtaining and interpreting the modality combining rules, in terms of the linear weights or odds-ratios on individual image modalities, three clinical applications are presented for prostate cancer segmentation, including modality availability assessment, importance quantification and rule discovery.
Active learning using adaptable task-based prioritisation
Dec 03, 2022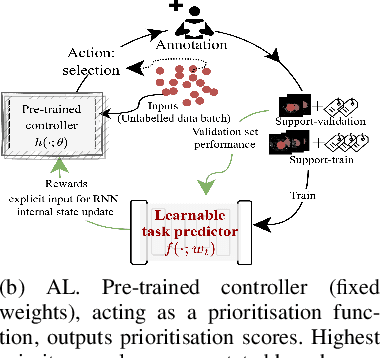
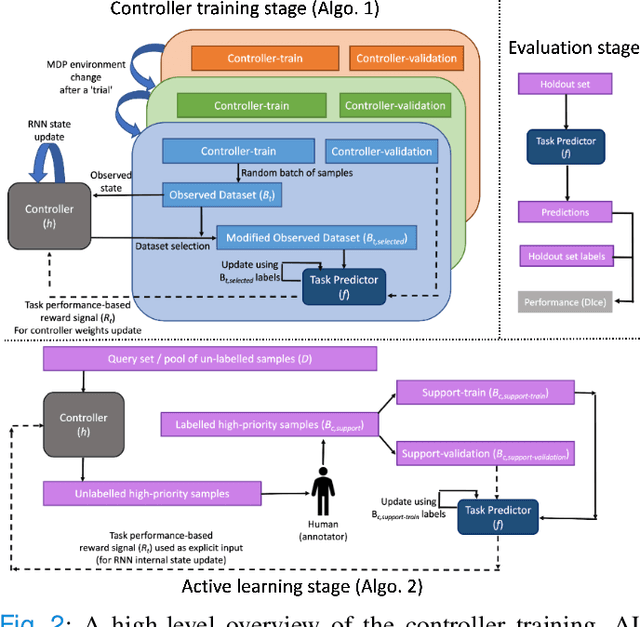
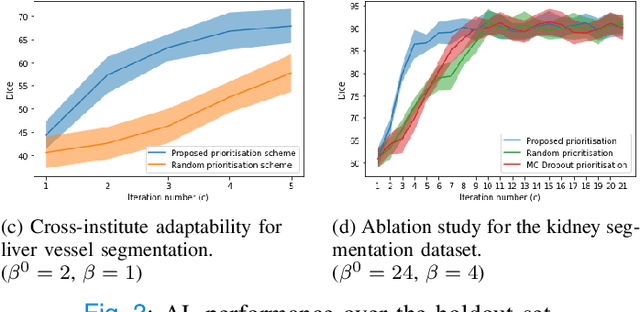
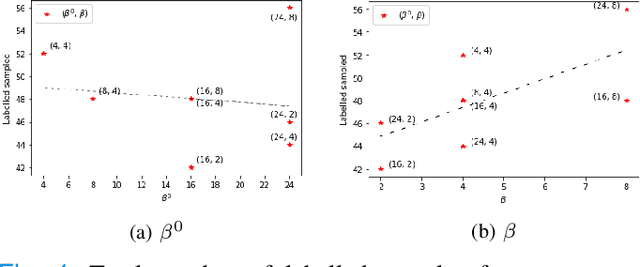
Abstract:Supervised machine learning-based medical image computing applications necessitate expert label curation, while unlabelled image data might be relatively abundant. Active learning methods aim to prioritise a subset of available image data for expert annotation, for label-efficient model training. We develop a controller neural network that measures priority of images in a sequence of batches, as in batch-mode active learning, for multi-class segmentation tasks. The controller is optimised by rewarding positive task-specific performance gain, within a Markov decision process (MDP) environment that also optimises the task predictor. In this work, the task predictor is a segmentation network. A meta-reinforcement learning algorithm is proposed with multiple MDPs, such that the pre-trained controller can be adapted to a new MDP that contains data from different institutes and/or requires segmentation of different organs or structures within the abdomen. We present experimental results using multiple CT datasets from more than one thousand patients, with segmentation tasks of nine different abdominal organs, to demonstrate the efficacy of the learnt prioritisation controller function and its cross-institute and cross-organ adaptability. We show that the proposed adaptable prioritisation metric yields converging segmentation accuracy for the novel class of kidney, unseen in training, using between approximately 40\% to 60\% of labels otherwise required with other heuristic or random prioritisation metrics. For clinical datasets of limited size, the proposed adaptable prioritisation offers a performance improvement of 22.6\% and 10.2\% in Dice score, for tasks of kidney and liver vessel segmentation, respectively, compared to random prioritisation and alternative active sampling strategies.
Trackerless freehand ultrasound with sequence modelling and auxiliary transformation over past and future frames
Nov 09, 2022



Abstract:Three-dimensional (3D) freehand ultrasound (US) reconstruction without a tracker can be advantageous over its two-dimensional or tracked counterparts in many clinical applications. In this paper, we propose to estimate 3D spatial transformation between US frames from both past and future 2D images, using feed-forward and recurrent neural networks (RNNs). With the temporally available frames, a further multi-task learning algorithm is proposed to utilise a large number of auxiliary transformation-predicting tasks between them. Using more than 40,000 US frames acquired from 228 scans on 38 forearms of 19 volunteers in a volunteer study, the hold-out test performance is quantified by frame prediction accuracy, volume reconstruction overlap, accumulated tracking error and final drift, based on ground-truth from an optical tracker. The results show the importance of modelling the temporal-spatially correlated input frames as well as output transformations, with further improvement owing to additional past and/or future frames. The best performing model was associated with predicting transformation between moderately-spaced frames, with an interval of less than ten frames at 20 frames per second (fps). Little benefit was observed by adding frames more than one second away from the predicted transformation, with or without LSTM-based RNNs. Interestingly, with the proposed approach, explicit within-sequence loss that encourages consistency in composing transformations or minimises accumulated error may no longer be required. The implementation code and volunteer data will be made publicly available ensuring reproducibility and further research.
Collaborative Quantization Embeddings for Intra-Subject Prostate MR Image Registration
Jul 14, 2022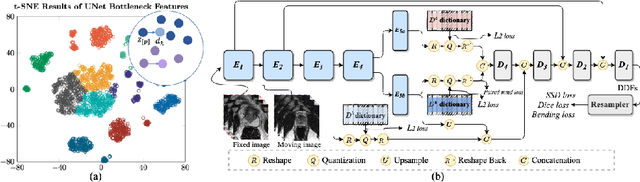
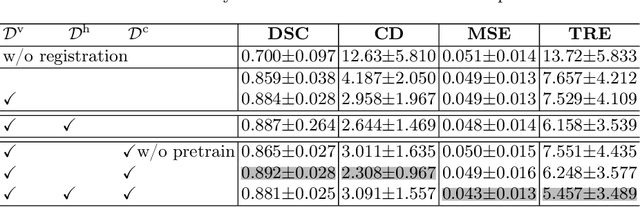
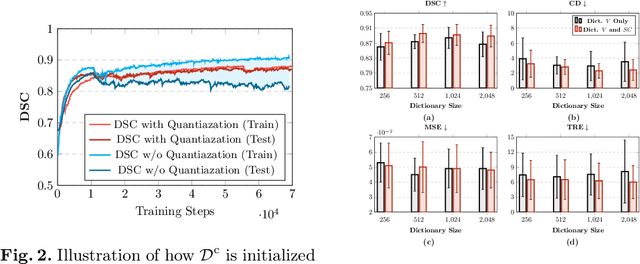
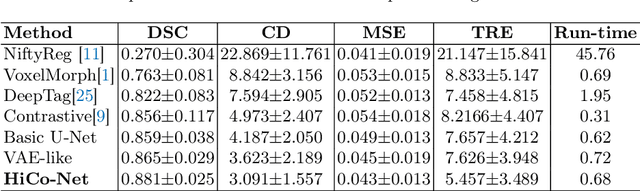
Abstract:Image registration is useful for quantifying morphological changes in longitudinal MR images from prostate cancer patients. This paper describes a development in improving the learning-based registration algorithms, for this challenging clinical application often with highly variable yet limited training data. First, we report that the latent space can be clustered into a much lower dimensional space than that commonly found as bottleneck features at the deep layer of a trained registration network. Based on this observation, we propose a hierarchical quantization method, discretizing the learned feature vectors using a jointly-trained dictionary with a constrained size, in order to improve the generalisation of the registration networks. Furthermore, a novel collaborative dictionary is independently optimised to incorporate additional prior information, such as the segmentation of the gland or other regions of interest, in the latent quantized space. Based on 216 real clinical images from 86 prostate cancer patients, we show the efficacy of both the designed components. Improved registration accuracy was obtained with statistical significance, in terms of both Dice on gland and target registration error on corresponding landmarks, the latter of which achieved 5.46 mm, an improvement of 28.7\% from the baseline without quantization. Experimental results also show that the difference in performance was indeed minimised between training and testing data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge