Mirabela Rusu
The role of self-supervised pretraining in differentially private medical image analysis
Jan 27, 2026Abstract:Differential privacy (DP) provides formal protection for sensitive data but typically incurs substantial losses in diagnostic performance. Model initialization has emerged as a critical factor in mitigating this degradation, yet the role of modern self-supervised learning under full-model DP remains poorly understood. Here, we present a large-scale evaluation of initialization strategies for differentially private medical image analysis, using chest radiograph classification as a representative benchmark with more than 800,000 images. Using state-of-the-art ConvNeXt models trained with DP-SGD across realistic privacy regimes, we compare non-domain-specific supervised ImageNet initialization, non-domain-specific self-supervised DINOv3 initialization, and domain-specific supervised pretraining on MIMIC-CXR, the largest publicly available chest radiograph dataset. Evaluations are conducted across five external datasets spanning diverse institutions and acquisition settings. We show that DINOv3 initialization consistently improves diagnostic utility relative to ImageNet initialization under DP, but remains inferior to domain-specific supervised pretraining, which achieves performance closest to non-private baselines. We further demonstrate that initialization choice strongly influences demographic fairness, cross-dataset generalization, and robustness to data scale and model capacity under privacy constraints. The results establish initialization strategy as a central determinant of utility, fairness, and generalization in differentially private medical imaging.
Agentic large language models improve retrieval-based radiology question answering
Aug 01, 2025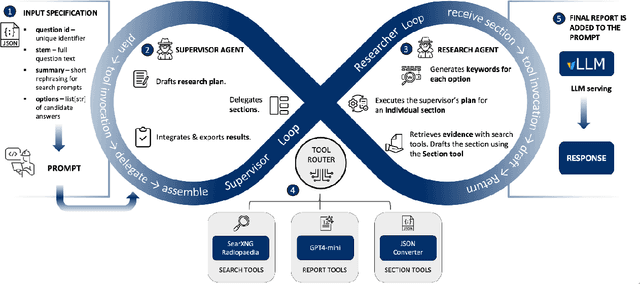
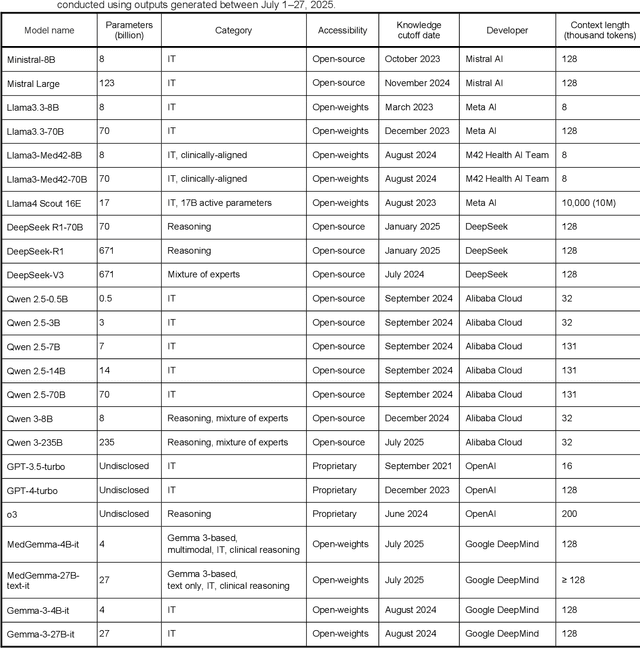
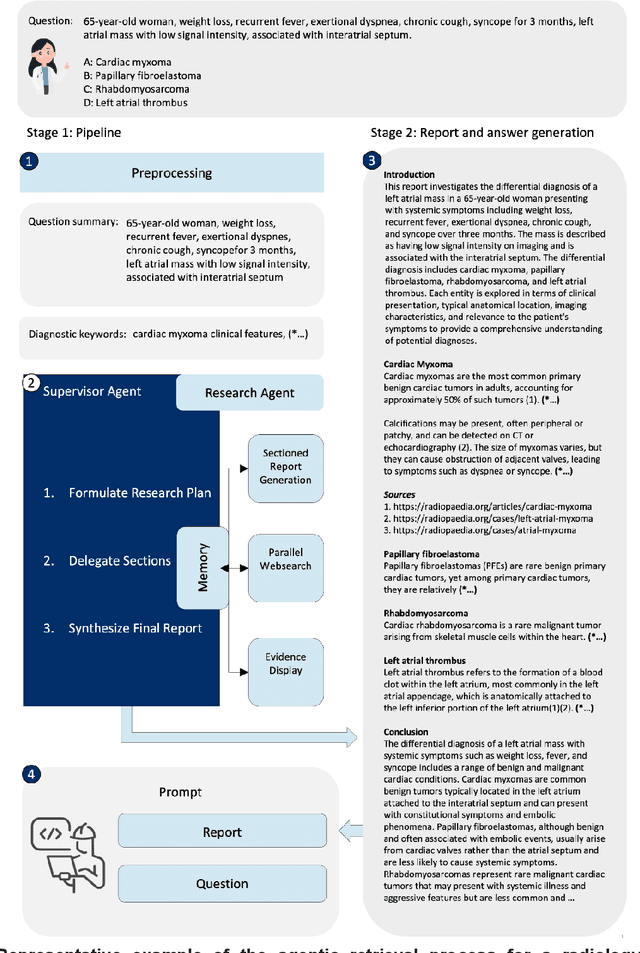
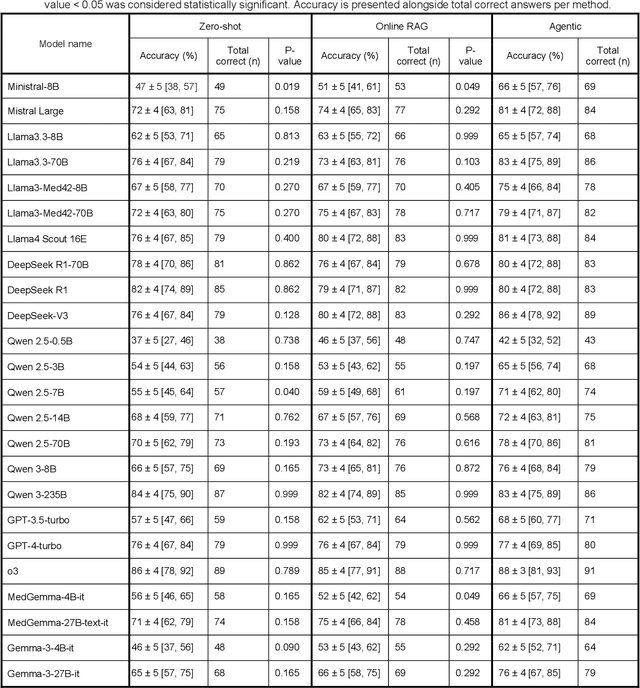
Abstract:Clinical decision-making in radiology increasingly benefits from artificial intelligence (AI), particularly through large language models (LLMs). However, traditional retrieval-augmented generation (RAG) systems for radiology question answering (QA) typically rely on single-step retrieval, limiting their ability to handle complex clinical reasoning tasks. Here we propose an agentic RAG framework enabling LLMs to autonomously decompose radiology questions, iteratively retrieve targeted clinical evidence from Radiopaedia, and dynamically synthesize evidence-based responses. We evaluated 24 LLMs spanning diverse architectures, parameter scales (0.5B to >670B), and training paradigms (general-purpose, reasoning-optimized, clinically fine-tuned), using 104 expert-curated radiology questions from previously established RSNA-RadioQA and ExtendedQA datasets. Agentic retrieval significantly improved mean diagnostic accuracy over zero-shot prompting (73% vs. 64%; P<0.001) and conventional online RAG (73% vs. 68%; P<0.001). The greatest gains occurred in mid-sized models (e.g., Mistral Large improved from 72% to 81%) and small-scale models (e.g., Qwen 2.5-7B improved from 55% to 71%), while very large models (>200B parameters) demonstrated minimal changes (<2% improvement). Additionally, agentic retrieval reduced hallucinations (mean 9.4%) and retrieved clinically relevant context in 46% of cases, substantially aiding factual grounding. Even clinically fine-tuned models exhibited meaningful improvements (e.g., MedGemma-27B improved from 71% to 81%), indicating complementary roles of retrieval and fine-tuning. These results highlight the potential of agentic frameworks to enhance factuality and diagnostic accuracy in radiology QA, particularly among mid-sized LLMs, warranting future studies to validate their clinical utility.
Promptable cancer segmentation using minimal expert-curated data
May 23, 2025Abstract:Automated segmentation of cancer on medical images can aid targeted diagnostic and therapeutic procedures. However, its adoption is limited by the high cost of expert annotations required for training and inter-observer variability in datasets. While weakly-supervised methods mitigate some challenges, using binary histology labels for training as opposed to requiring full segmentation, they require large paired datasets of histology and images, which are difficult to curate. Similarly, promptable segmentation aims to allow segmentation with no re-training for new tasks at inference, however, existing models perform poorly on pathological regions, again necessitating large datasets for training. In this work we propose a novel approach for promptable segmentation requiring only 24 fully-segmented images, supplemented by 8 weakly-labelled images, for training. Curating this minimal data to a high standard is relatively feasible and thus issues with the cost and variability of obtaining labels can be mitigated. By leveraging two classifiers, one weakly-supervised and one fully-supervised, our method refines segmentation through a guided search process initiated by a single-point prompt. Our approach outperforms existing promptable segmentation methods, and performs comparably with fully-supervised methods, for the task of prostate cancer segmentation, while using substantially less annotated data (up to 100X less). This enables promptable segmentation with very minimal labelled data, such that the labels can be curated to a very high standard.
Registration-Enhanced Segmentation Method for Prostate Cancer in Ultrasound Images
Feb 02, 2025



Abstract:Prostate cancer is a major cause of cancer-related deaths in men, where early detection greatly improves survival rates. Although MRI-TRUS fusion biopsy offers superior accuracy by combining MRI's detailed visualization with TRUS's real-time guidance, it is a complex and time-intensive procedure that relies heavily on manual annotations, leading to potential errors. To address these challenges, we propose a fully automatic MRI-TRUS fusion-based segmentation method that identifies prostate tumors directly in TRUS images without requiring manual annotations. Unlike traditional multimodal fusion approaches that rely on naive data concatenation, our method integrates a registration-segmentation framework to align and leverage spatial information between MRI and TRUS modalities. This alignment enhances segmentation accuracy and reduces reliance on manual effort. Our approach was validated on a dataset of 1,747 patients from Stanford Hospital, achieving an average Dice coefficient of 0.212, outperforming TRUS-only (0.117) and naive MRI-TRUS fusion (0.132) methods, with significant improvements (p $<$ 0.01). This framework demonstrates the potential for reducing the complexity of prostate cancer diagnosis and provides a flexible architecture applicable to other multimodal medical imaging tasks.
Mask Enhanced Deeply Supervised Prostate Cancer Detection on B-mode Micro-Ultrasound
Dec 14, 2024



Abstract:Prostate cancer is a leading cause of cancer-related deaths among men. The recent development of high frequency, micro-ultrasound imaging offers improved resolution compared to conventional ultrasound and potentially a better ability to differentiate clinically significant cancer from normal tissue. However, the features of prostate cancer remain subtle, with ambiguous borders with normal tissue and large variations in appearance, making it challenging for both machine learning and humans to localize it on micro-ultrasound images. We propose a novel Mask Enhanced Deeply-supervised Micro-US network, termed MedMusNet, to automatically and more accurately segment prostate cancer to be used as potential targets for biopsy procedures. MedMusNet leverages predicted masks of prostate cancer to enforce the learned features layer-wisely within the network, reducing the influence of noise and improving overall consistency across frames. MedMusNet successfully detected 76% of clinically significant cancer with a Dice Similarity Coefficient of 0.365, significantly outperforming the baseline Swin-M2F in specificity and accuracy (Wilcoxon test, Bonferroni correction, p-value<0.05). While the lesion-level and patient-level analyses showed improved performance compared to human experts and different baseline, the improvements did not reach statistical significance, likely on account of the small cohort. We have presented a novel approach to automatically detect and segment clinically significant prostate cancer on B-mode micro-ultrasound images. Our MedMusNet model outperformed other models, surpassing even human experts. These preliminary results suggest the potential for aiding urologists in prostate cancer diagnosis via biopsy and treatment decision-making.
BreastRegNet: A Deep Learning Framework for Registration of Breast Faxitron and Histopathology Images
Jan 18, 2024Abstract:A standard treatment protocol for breast cancer entails administering neoadjuvant therapy followed by surgical removal of the tumor and surrounding tissue. Pathologists typically rely on cabinet X-ray radiographs, known as Faxitron, to examine the excised breast tissue and diagnose the extent of residual disease. However, accurately determining the location, size, and focality of residual cancer can be challenging, and incorrect assessments can lead to clinical consequences. The utilization of automated methods can improve the histopathology process, allowing pathologists to choose regions for sampling more effectively and precisely. Despite the recognized necessity, there are currently no such methods available. Training such automated detection models require accurate ground truth labels on ex-vivo radiology images, which can be acquired through registering Faxitron and histopathology images and mapping the extent of cancer from histopathology to x-ray images. This study introduces a deep learning-based image registration approach trained on mono-modal synthetic image pairs. The models were trained using data from 50 women who received neoadjuvant chemotherapy and underwent surgery. The results demonstrate that our method is faster and yields significantly lower average landmark error ($2.1\pm1.96$ mm) over the state-of-the-art iterative ($4.43\pm4.1$ mm) and deep learning ($4.02\pm3.15$ mm) approaches. Improved performance of our approach in integrating radiology and pathology information facilitates generating large datasets, which allows training models for more accurate breast cancer detection.
ProsDectNet: Bridging the Gap in Prostate Cancer Detection via Transrectal B-mode Ultrasound Imaging
Dec 08, 2023



Abstract:Interpreting traditional B-mode ultrasound images can be challenging due to image artifacts (e.g., shadowing, speckle), leading to low sensitivity and limited diagnostic accuracy. While Magnetic Resonance Imaging (MRI) has been proposed as a solution, it is expensive and not widely available. Furthermore, most biopsies are guided by Transrectal Ultrasound (TRUS) alone and can miss up to 52% cancers, highlighting the need for improved targeting. To address this issue, we propose ProsDectNet, a multi-task deep learning approach that localizes prostate cancer on B-mode ultrasound. Our model is pre-trained using radiologist-labeled data and fine-tuned using biopsy-confirmed labels. ProsDectNet includes a lesion detection and patch classification head, with uncertainty minimization using entropy to improve model performance and reduce false positive predictions. We trained and validated ProsDectNet using a cohort of 289 patients who underwent MRI-TRUS fusion targeted biopsy. We then tested our approach on a group of 41 patients and found that ProsDectNet outperformed the average expert clinician in detecting prostate cancer on B-mode ultrasound images, achieving a patient-level ROC-AUC of 82%, a sensitivity of 74%, and a specificity of 67%. Our results demonstrate that ProsDectNet has the potential to be used as a computer-aided diagnosis system to improve targeted biopsy and treatment planning.
Correlated Feature Aggregation by Region Helps Distinguish Aggressive from Indolent Clear Cell Renal Cell Carcinoma Subtypes on CT
Sep 29, 2022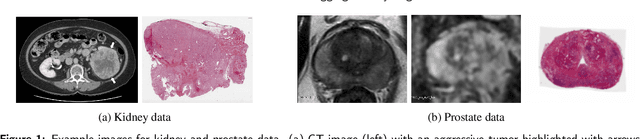
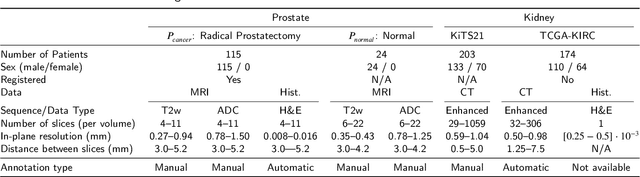
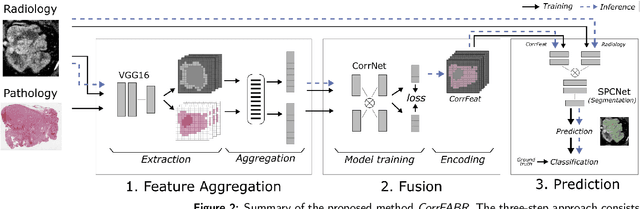

Abstract:Renal cell carcinoma (RCC) is a common cancer that varies in clinical behavior. Indolent RCC is often low-grade without necrosis and can be monitored without treatment. Aggressive RCC is often high-grade and can cause metastasis and death if not promptly detected and treated. While most kidney cancers are detected on CT scans, grading is based on histology from invasive biopsy or surgery. Determining aggressiveness on CT images is clinically important as it facilitates risk stratification and treatment planning. This study aims to use machine learning methods to identify radiology features that correlate with features on pathology to facilitate assessment of cancer aggressiveness on CT images instead of histology. This paper presents a novel automated method, Correlated Feature Aggregation By Region (CorrFABR), for classifying aggressiveness of clear cell RCC by leveraging correlations between radiology and corresponding unaligned pathology images. CorrFABR consists of three main steps: (1) Feature Aggregation where region-level features are extracted from radiology and pathology images, (2) Fusion where radiology features correlated with pathology features are learned on a region level, and (3) Prediction where the learned correlated features are used to distinguish aggressive from indolent clear cell RCC using CT alone as input. Thus, during training, CorrFABR learns from both radiology and pathology images, but during inference, CorrFABR will distinguish aggressive from indolent clear cell RCC using CT alone, in the absence of pathology images. CorrFABR improved classification performance over radiology features alone, with an increase in binary classification F1-score from 0.68 (0.04) to 0.73 (0.03). This demonstrates the potential of incorporating pathology disease characteristics for improved classification of aggressiveness of clear cell RCC on CT images.
Domain Generalization for Prostate Segmentation in Transrectal Ultrasound Images: A Multi-center Study
Sep 05, 2022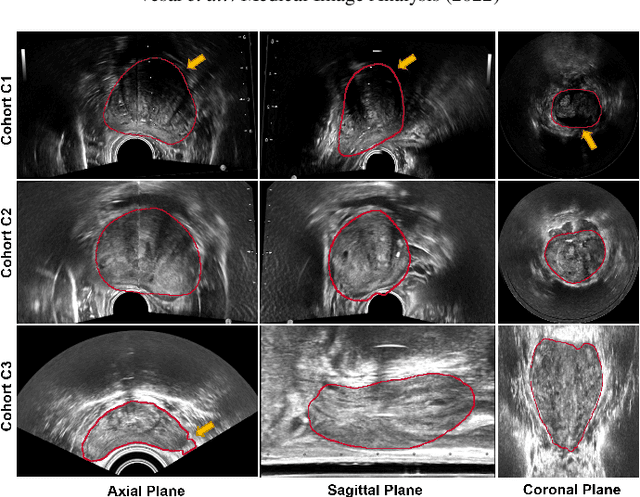
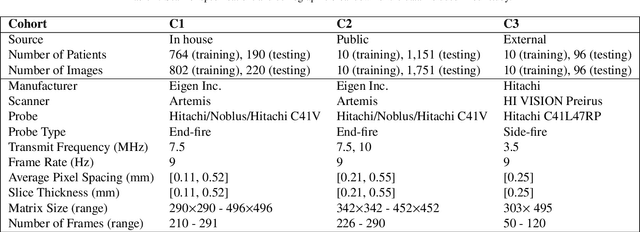
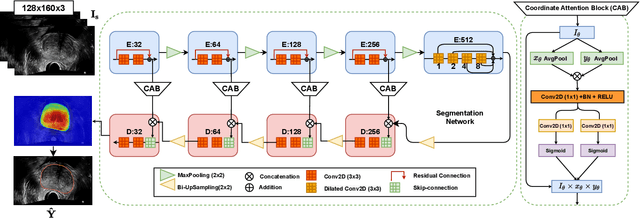
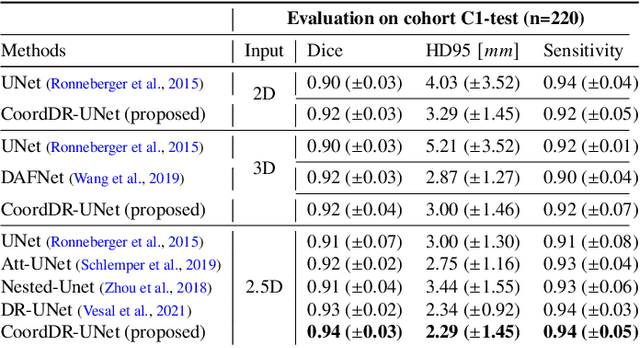
Abstract:Prostate biopsy and image-guided treatment procedures are often performed under the guidance of ultrasound fused with magnetic resonance images (MRI). Accurate image fusion relies on accurate segmentation of the prostate on ultrasound images. Yet, the reduced signal-to-noise ratio and artifacts (e.g., speckle and shadowing) in ultrasound images limit the performance of automated prostate segmentation techniques and generalizing these methods to new image domains is inherently difficult. In this study, we address these challenges by introducing a novel 2.5D deep neural network for prostate segmentation on ultrasound images. Our approach addresses the limitations of transfer learning and finetuning methods (i.e., drop in performance on the original training data when the model weights are updated) by combining a supervised domain adaptation technique and a knowledge distillation loss. The knowledge distillation loss allows the preservation of previously learned knowledge and reduces the performance drop after model finetuning on new datasets. Furthermore, our approach relies on an attention module that considers model feature positioning information to improve the segmentation accuracy. We trained our model on 764 subjects from one institution and finetuned our model using only ten subjects from subsequent institutions. We analyzed the performance of our method on three large datasets encompassing 2067 subjects from three different institutions. Our method achieved an average Dice Similarity Coefficient (Dice) of $94.0\pm0.03$ and Hausdorff Distance (HD95) of 2.28 $mm$ in an independent set of subjects from the first institution. Moreover, our model generalized well in the studies from the other two institutions (Dice: $91.0\pm0.03$; HD95: 3.7$mm$ and Dice: $82.0\pm0.03$; HD95: 7.1 $mm$).
Collaborative Quantization Embeddings for Intra-Subject Prostate MR Image Registration
Jul 14, 2022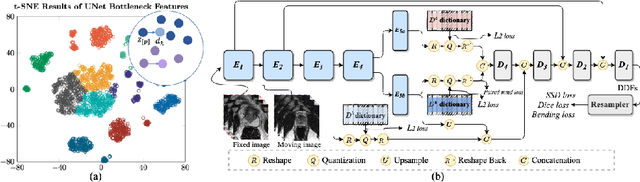
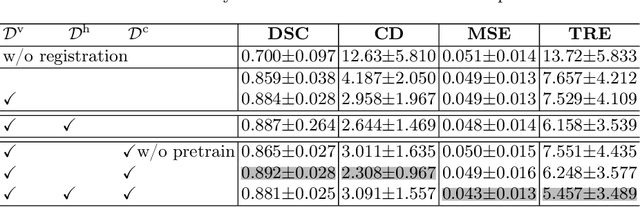
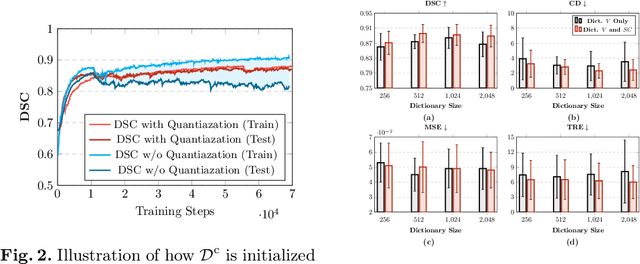
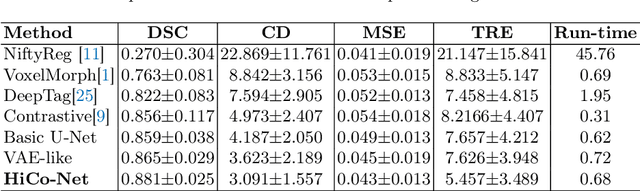
Abstract:Image registration is useful for quantifying morphological changes in longitudinal MR images from prostate cancer patients. This paper describes a development in improving the learning-based registration algorithms, for this challenging clinical application often with highly variable yet limited training data. First, we report that the latent space can be clustered into a much lower dimensional space than that commonly found as bottleneck features at the deep layer of a trained registration network. Based on this observation, we propose a hierarchical quantization method, discretizing the learned feature vectors using a jointly-trained dictionary with a constrained size, in order to improve the generalisation of the registration networks. Furthermore, a novel collaborative dictionary is independently optimised to incorporate additional prior information, such as the segmentation of the gland or other regions of interest, in the latent quantized space. Based on 216 real clinical images from 86 prostate cancer patients, we show the efficacy of both the designed components. Improved registration accuracy was obtained with statistical significance, in terms of both Dice on gland and target registration error on corresponding landmarks, the latter of which achieved 5.46 mm, an improvement of 28.7\% from the baseline without quantization. Experimental results also show that the difference in performance was indeed minimised between training and testing data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge