Geoffrey Sonn
On Aligning Prediction Models with Clinical Experiential Learning: A Prostate Cancer Case Study
Sep 04, 2025



Abstract:Over the past decade, the use of machine learning (ML) models in healthcare applications has rapidly increased. Despite high performance, modern ML models do not always capture patterns the end user requires. For example, a model may predict a non-monotonically decreasing relationship between cancer stage and survival, keeping all other features fixed. In this paper, we present a reproducible framework for investigating this misalignment between model behavior and clinical experiential learning, focusing on the effects of underspecification of modern ML pipelines. In a prostate cancer outcome prediction case study, we first identify and address these inconsistencies by incorporating clinical knowledge, collected by a survey, via constraints into the ML model, and subsequently analyze the impact on model performance and behavior across degrees of underspecification. The approach shows that aligning the ML model with clinical experiential learning is possible without compromising performance. Motivated by recent literature in generative AI, we further examine the feasibility of a feedback-driven alignment approach in non-generative AI clinical risk prediction models through a randomized experiment with clinicians. Our findings illustrate that, by eliciting clinicians' model preferences using our proposed methodology, the larger the difference in how the constrained and unconstrained models make predictions for a patient, the more apparent the difference is in clinical interpretation.
Using Fiber Optic Bundles to Miniaturize Vision-Based Tactile Sensors
Mar 12, 2024



Abstract:Vision-based tactile sensors have recently become popular due to their combination of low cost, very high spatial resolution, and ease of integration using widely available miniature cameras. The associated field of view and focal length, however, are difficult to package in a human-sized finger. In this paper we employ optical fiber bundles to achieve a form factor that, at 15 mm diameter, is smaller than an average human fingertip. The electronics and camera are also located remotely, further reducing package size. The sensor achieves a spatial resolution of 0.22 mm and a minimum force resolution 5 mN for normal and shear contact forces. With these attributes, the DIGIT Pinki sensor is suitable for applications such as robotic and teleoperated digital palpation. We demonstrate its utility for palpation of the prostate gland and show that it can achieve clinically relevant discrimination of prostate stiffness for phantom and ex vivo tissue.
Collaborative Quantization Embeddings for Intra-Subject Prostate MR Image Registration
Jul 14, 2022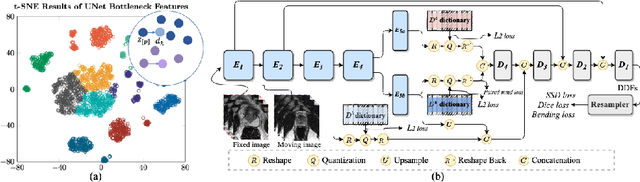
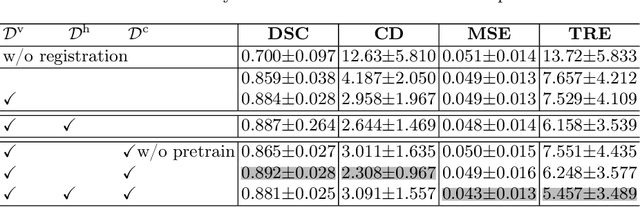
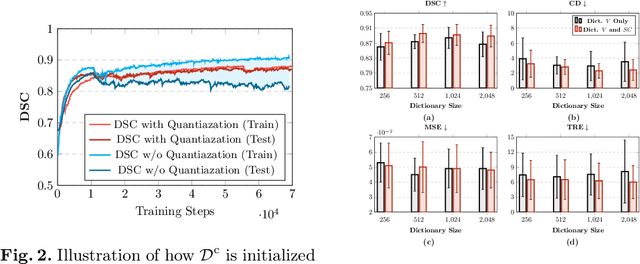
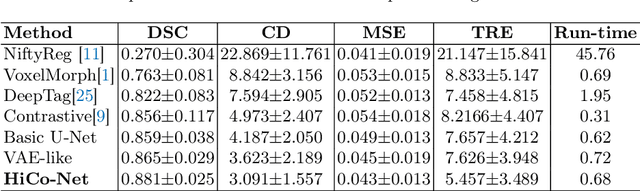
Abstract:Image registration is useful for quantifying morphological changes in longitudinal MR images from prostate cancer patients. This paper describes a development in improving the learning-based registration algorithms, for this challenging clinical application often with highly variable yet limited training data. First, we report that the latent space can be clustered into a much lower dimensional space than that commonly found as bottleneck features at the deep layer of a trained registration network. Based on this observation, we propose a hierarchical quantization method, discretizing the learned feature vectors using a jointly-trained dictionary with a constrained size, in order to improve the generalisation of the registration networks. Furthermore, a novel collaborative dictionary is independently optimised to incorporate additional prior information, such as the segmentation of the gland or other regions of interest, in the latent quantized space. Based on 216 real clinical images from 86 prostate cancer patients, we show the efficacy of both the designed components. Improved registration accuracy was obtained with statistical significance, in terms of both Dice on gland and target registration error on corresponding landmarks, the latter of which achieved 5.46 mm, an improvement of 28.7\% from the baseline without quantization. Experimental results also show that the difference in performance was indeed minimised between training and testing data.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021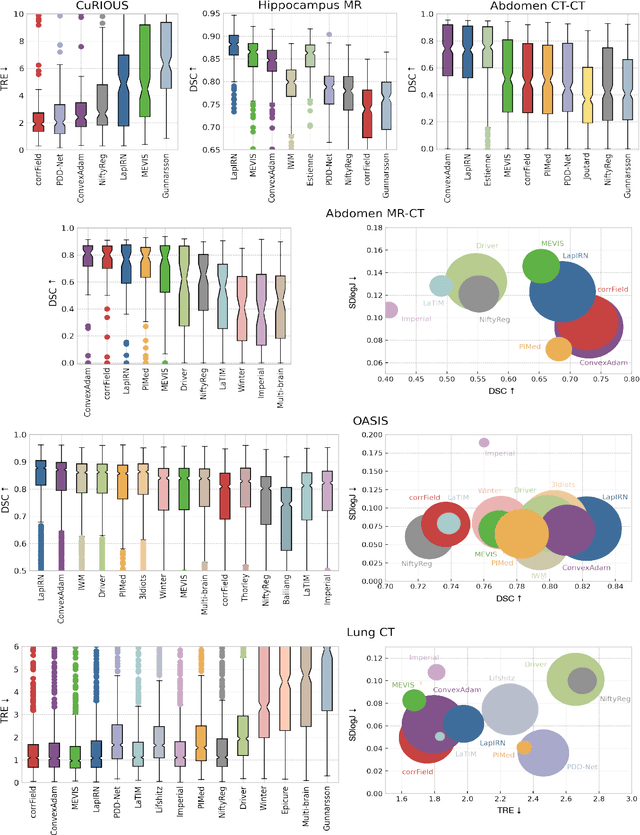
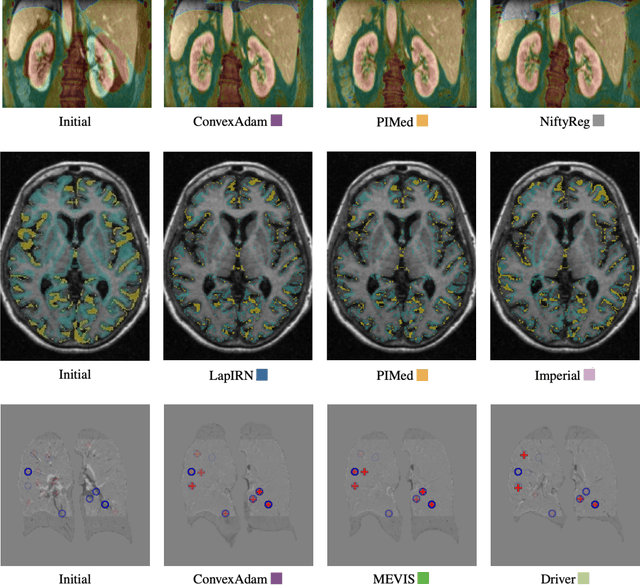
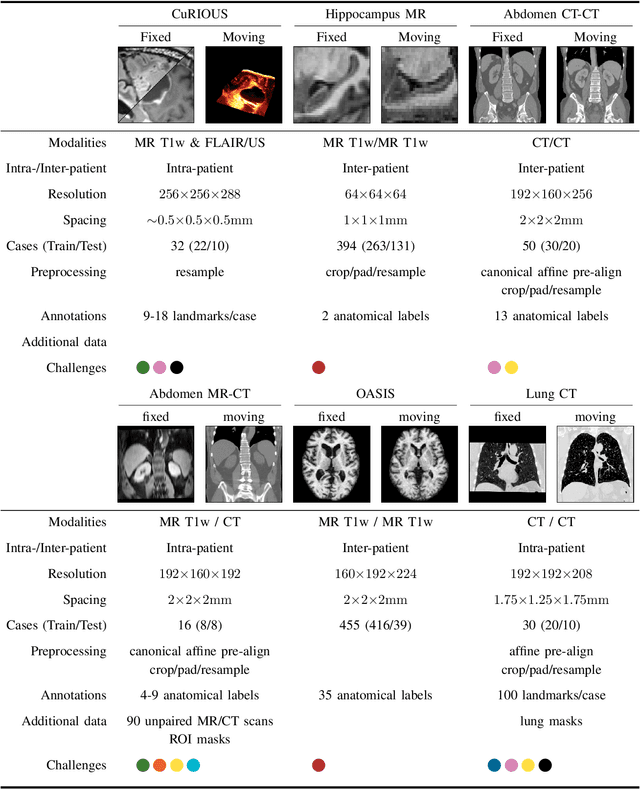
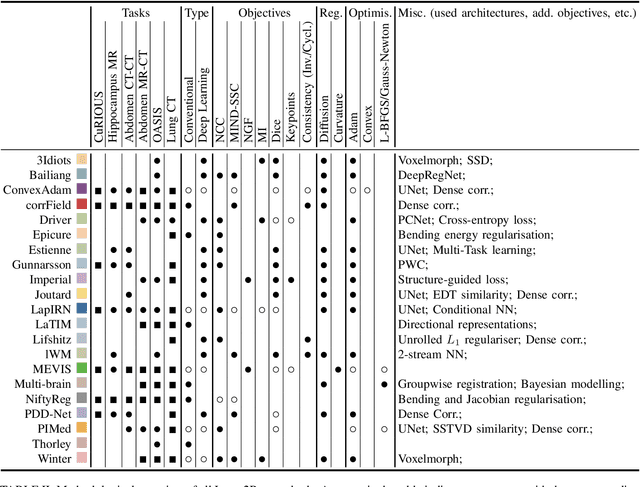
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
Deep Learning for Prostate Pathology
Oct 16, 2019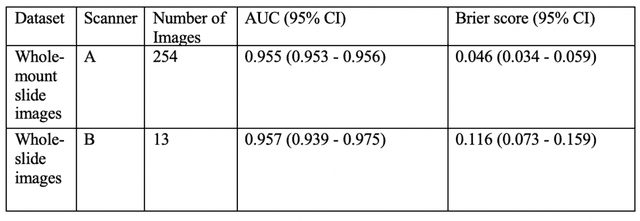
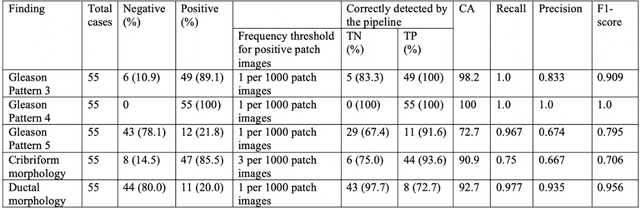
Abstract:The current study detects different morphologies related to prostate pathology using deep learning models; these models were evaluated on 2,121 hematoxylin and eosin (H&E) stain histology images captured using bright field microscopy, which spanned a variety of image qualities, origins (whole slide, tissue micro array, whole mount, Internet), scanning machines, timestamps, H&E staining protocols, and institutions. For case usage, these models were applied for the annotation tasks in clinician-oriented pathology reports for prostatectomy specimens. The true positive rate (TPR) for slides with prostate cancer was 99.7% by a false positive rate of 0.785%. The F1-scores of Gleason patterns reported in pathology reports ranged from 0.795 to 1.0 at the case level. TPR was 93.6% for the cribriform morphology and 72.6% for the ductal morphology. The correlation between the ground truth and the prediction for the relative tumor volume was 0.987 n. Our models cover the major components of prostate pathology and successfully accomplish the annotation tasks.
Computerized Multiparametric MR image Analysis for Prostate Cancer Aggressiveness-Assessment
Dec 01, 2016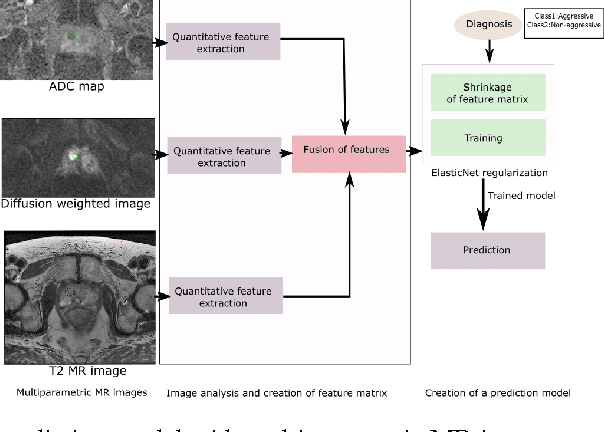
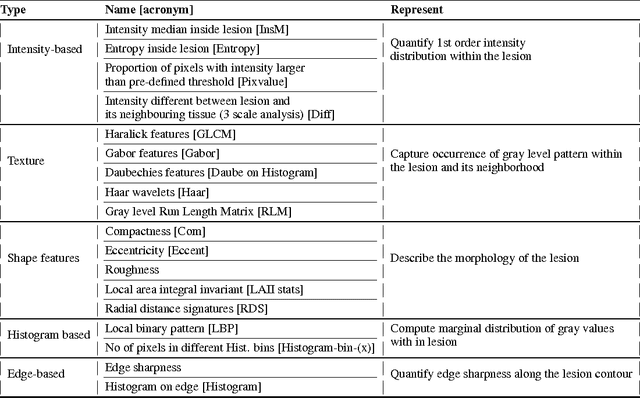
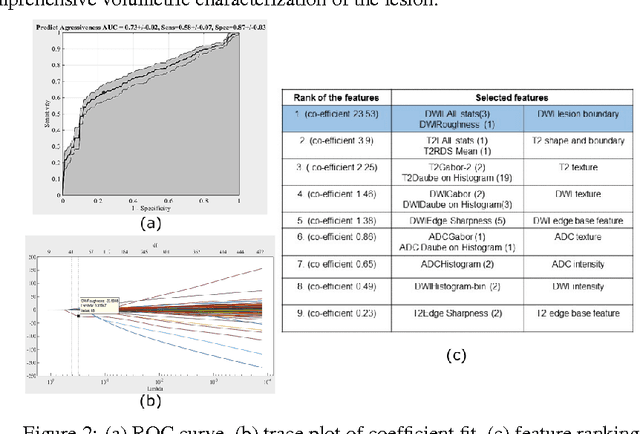
Abstract:We propose an automated method for detecting aggressive prostate cancer(CaP) (Gleason score >=7) based on a comprehensive analysis of the lesion and the surrounding normal prostate tissue which has been simultaneously captured in T2-weighted MR images, diffusion-weighted images (DWI) and apparent diffusion coefficient maps (ADC). The proposed methodology was tested on a dataset of 79 patients (40 aggressive, 39 non-aggressive). We evaluated the performance of a wide range of popular quantitative imaging features on the characterization of aggressive versus non-aggressive CaP. We found that a group of 44 discriminative predictors among 1464 quantitative imaging features can be used to produce an area under the ROC curve of 0.73.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge