Andreas Loening
Biologic and Prognostic Feature Scores from Whole-Slide Histology Images Using Deep Learning
Oct 24, 2019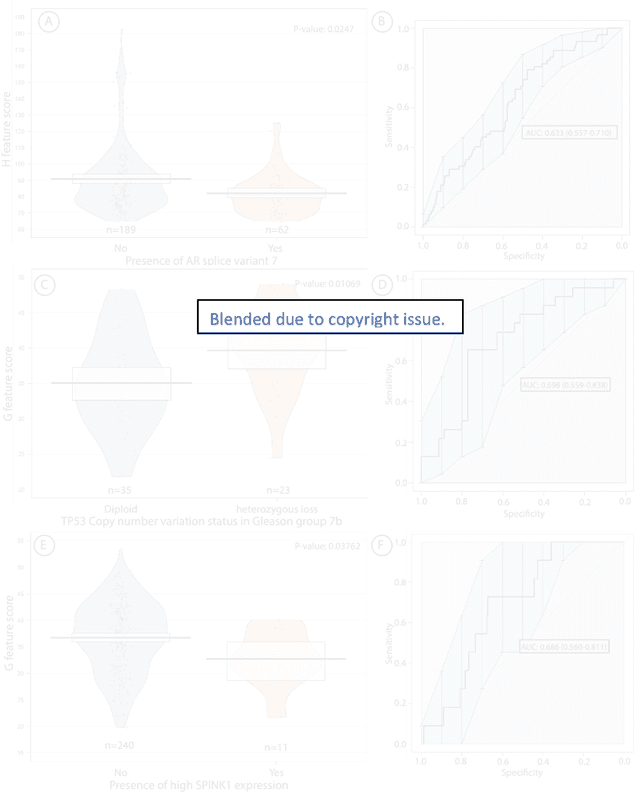
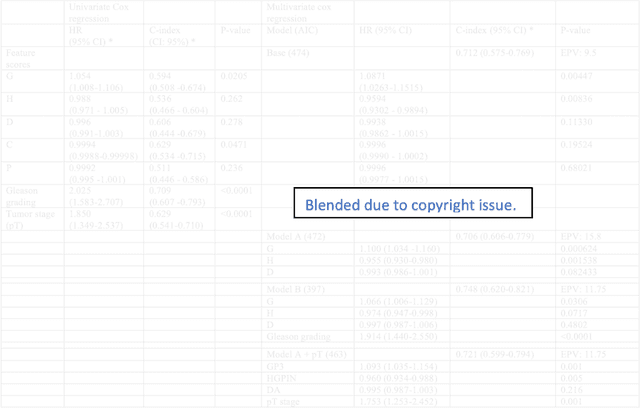
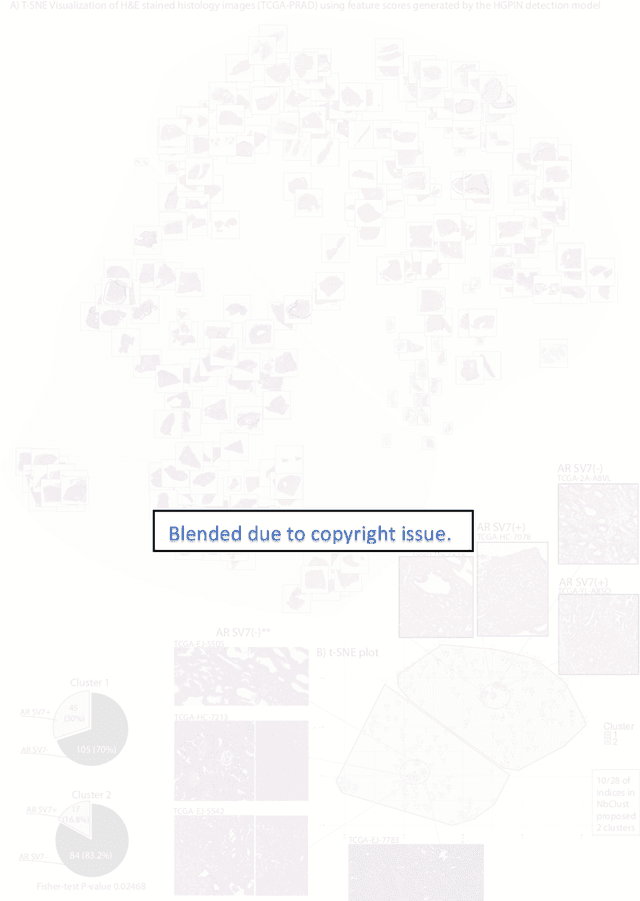
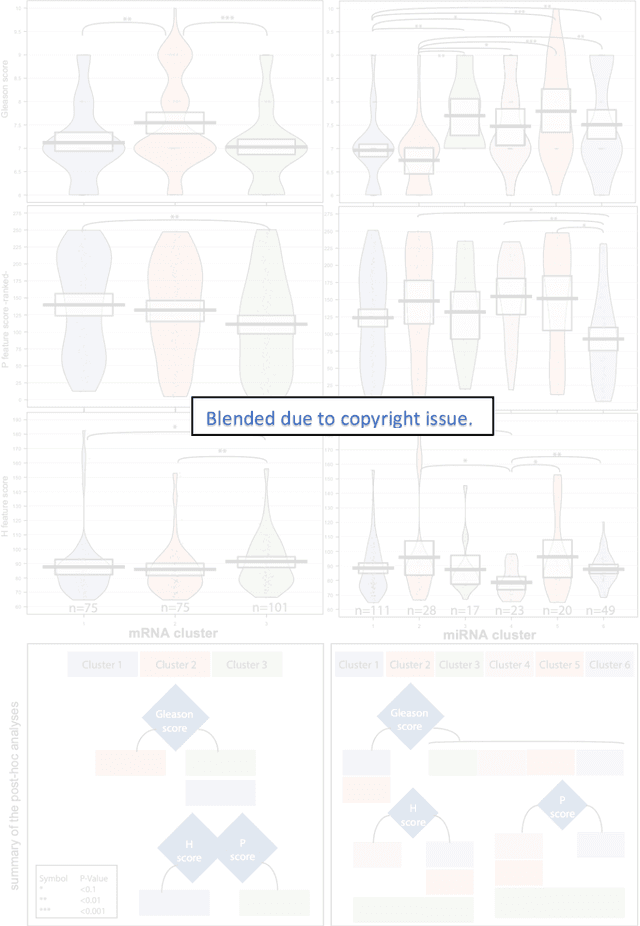
Abstract:Histopathology is a reflection of the molecular changes and provides prognostic phenotypes representing the disease progression. In this study, we introduced feature scores generated from hematoxylin and eosin histology images based on deep learning (DL) models developed for prostate pathology. We demonstrated that these feature scores were significantly prognostic for time to event endpoints (biochemical recurrence and cancer-specific survival) and had simultaneously molecular biologic associations to relevant genomic alterations and molecular subtypes using already trained DL models that were not previously exposed to the datasets of the current study. Further, we discussed the potential of such feature scores to improve the current tumor grading system and the challenges that are associated with tumor heterogeneity and the development of prognostic models from histology images. Our findings uncover the potential of feature scores from histology images as digital biomarkers in precision medicine and as an expanding utility for digital pathology.
Deep Learning for Prostate Pathology
Oct 16, 2019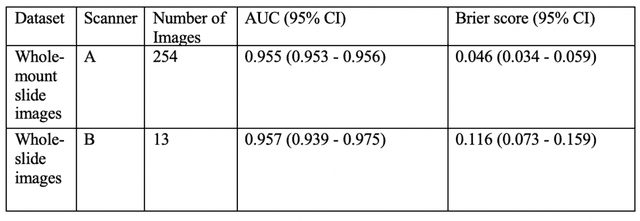
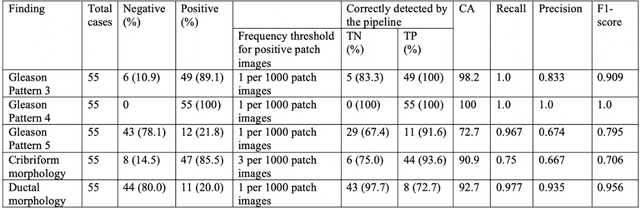
Abstract:The current study detects different morphologies related to prostate pathology using deep learning models; these models were evaluated on 2,121 hematoxylin and eosin (H&E) stain histology images captured using bright field microscopy, which spanned a variety of image qualities, origins (whole slide, tissue micro array, whole mount, Internet), scanning machines, timestamps, H&E staining protocols, and institutions. For case usage, these models were applied for the annotation tasks in clinician-oriented pathology reports for prostatectomy specimens. The true positive rate (TPR) for slides with prostate cancer was 99.7% by a false positive rate of 0.785%. The F1-scores of Gleason patterns reported in pathology reports ranged from 0.795 to 1.0 at the case level. TPR was 93.6% for the cribriform morphology and 72.6% for the ductal morphology. The correlation between the ground truth and the prediction for the relative tumor volume was 0.987 n. Our models cover the major components of prostate pathology and successfully accomplish the annotation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge