Yuri Tolkach
Pathologist-like explainable AI for interpretable Gleason grading in prostate cancer
Oct 19, 2024



Abstract:The aggressiveness of prostate cancer, the most common cancer in men worldwide, is primarily assessed based on histopathological data using the Gleason scoring system. While artificial intelligence (AI) has shown promise in accurately predicting Gleason scores, these predictions often lack inherent explainability, potentially leading to distrust in human-machine interactions. To address this issue, we introduce a novel dataset of 1,015 tissue microarray core images, annotated by an international group of 54 pathologists. The annotations provide detailed localized pattern descriptions for Gleason grading in line with international guidelines. Utilizing this dataset, we develop an inherently explainable AI system based on a U-Net architecture that provides predictions leveraging pathologists' terminology. This approach circumvents post-hoc explainability methods while maintaining or exceeding the performance of methods trained directly for Gleason pattern segmentation (Dice score: 0.713 $\pm$ 0.003 trained on explanations vs. 0.691 $\pm$ 0.010 trained on Gleason patterns). By employing soft labels during training, we capture the intrinsic uncertainty in the data, yielding strong results in Gleason pattern segmentation even in the context of high interobserver variability. With the release of this dataset, we aim to encourage further research into segmentation in medical tasks with high levels of subjectivity and to advance the understanding of pathologists' reasoning processes.
From Pointwise to Powerhouse: Initialising Neural Networks with Generative Models
Oct 25, 2023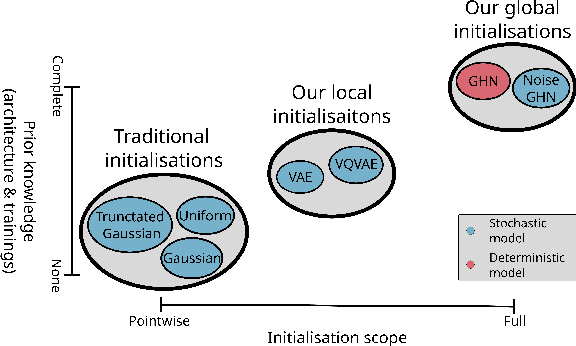
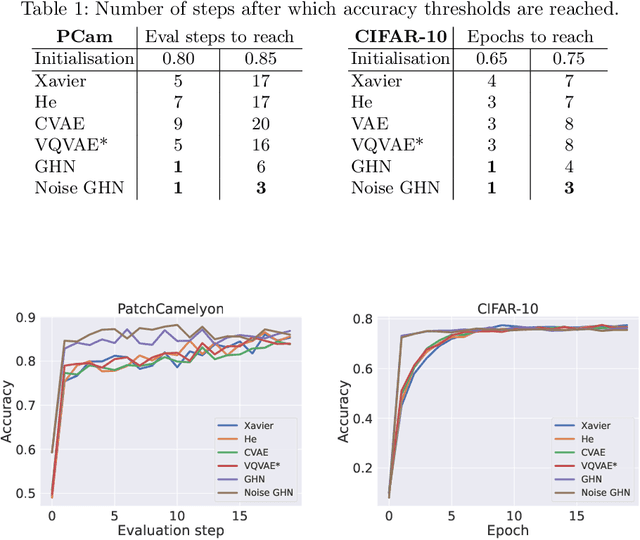
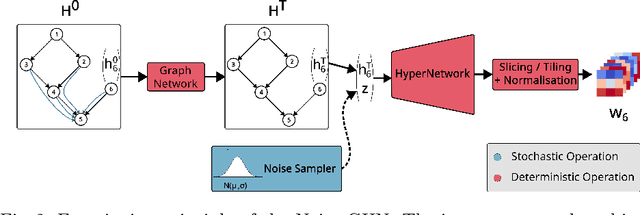
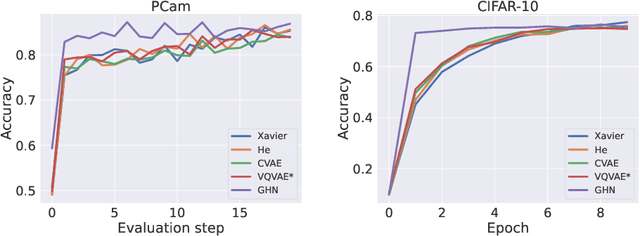
Abstract:Traditional initialisation methods, e.g. He and Xavier, have been effective in avoiding the problem of vanishing or exploding gradients in neural networks. However, they only use simple pointwise distributions, which model one-dimensional variables. Moreover, they ignore most information about the architecture and disregard past training experiences. These limitations can be overcome by employing generative models for initialisation. In this paper, we introduce two groups of new initialisation methods. First, we locally initialise weight groups by employing variational autoencoders. Secondly, we globally initialise full weight sets by employing graph hypernetworks. We thoroughly evaluate the impact of the employed generative models on state-of-the-art neural networks in terms of accuracy, convergence speed and ensembling. Our results show that global initialisations result in higher accuracy and faster initial convergence speed. However, the implementation through graph hypernetworks leads to diminished ensemble performance on out of distribution data. To counteract, we propose a modification called noise graph hypernetwork, which encourages diversity in the produced ensemble members. Furthermore, our approach might be able to transfer learned knowledge to different image distributions. Our work provides insights into the potential, the trade-offs and possible modifications of these new initialisation methods.
Jointly Exploring Client Drift and Catastrophic Forgetting in Dynamic Learning
Sep 01, 2023



Abstract:Federated and Continual Learning have emerged as potential paradigms for the robust and privacy-aware use of Deep Learning in dynamic environments. However, Client Drift and Catastrophic Forgetting are fundamental obstacles to guaranteeing consistent performance. Existing work only addresses these problems separately, which neglects the fact that the root cause behind both forms of performance deterioration is connected. We propose a unified analysis framework for building a controlled test environment for Client Drift -- by perturbing a defined ratio of clients -- and Catastrophic Forgetting -- by shifting all clients with a particular strength. Our framework further leverages this new combined analysis by generating a 3D landscape of the combined performance impact from both. We demonstrate that the performance drop through Client Drift, caused by a certain share of shifted clients, is correlated to the drop from Catastrophic Forgetting resulting from a corresponding shift strength. Correlation tests between both problems for Computer Vision (CelebA) and Medical Imaging (PESO) support this new perspective, with an average Pearson rank correlation coefficient of over 0.94. Our framework's novel ability of combined spatio-temporal shift analysis allows us to investigate how both forms of distribution shift behave in mixed scenarios, opening a new pathway for better generalization. We show that a combination of moderate Client Drift and Catastrophic Forgetting can even improve the performance of the resulting model (causing a "Generalization Bump") compared to when only one of the shifts occurs individually. We apply a simple and commonly used method from Continual Learning in the federated setting and observe this phenomenon to be reoccurring, leveraging the ability of our framework to analyze existing and novel methods for Federated and Continual Learning.
Critical Evaluation of Artificial Intelligence as Digital Twin of Pathologist for Prostate Cancer Pathology
Aug 23, 2023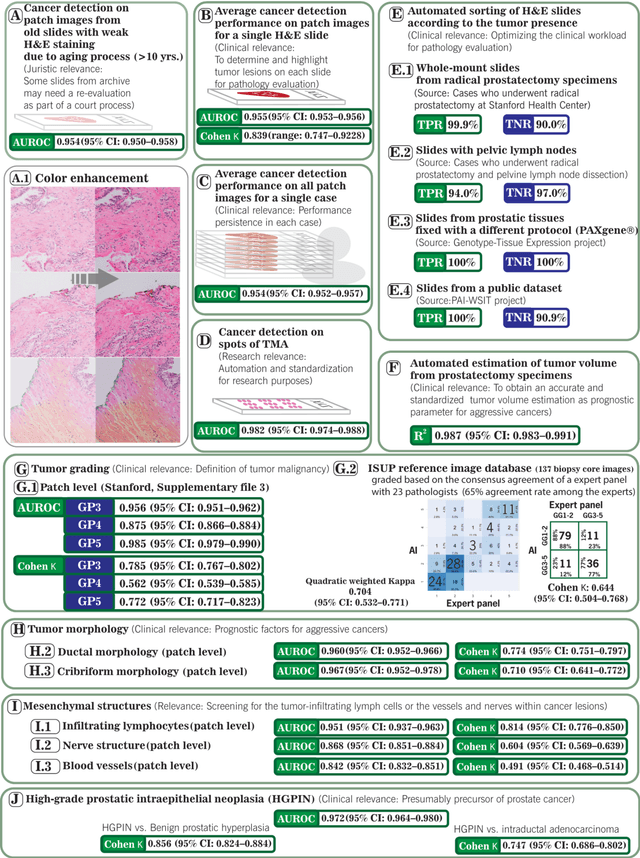
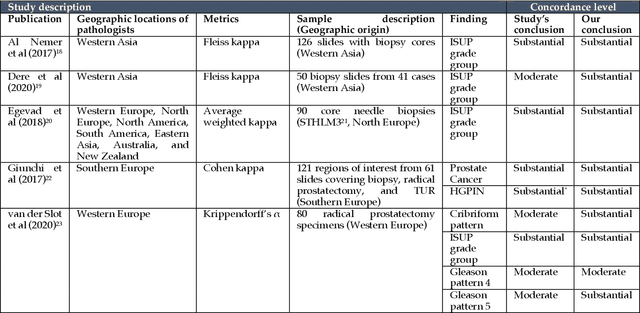
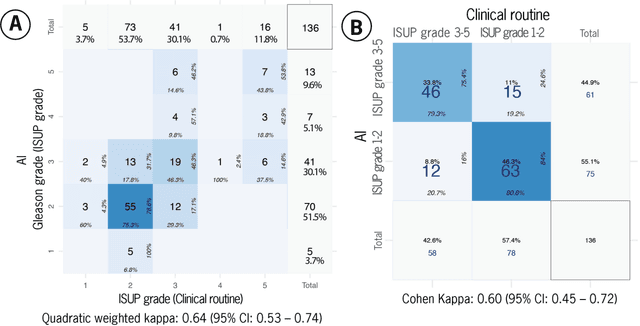
Abstract:Prostate cancer pathology plays a crucial role in clinical management but is time-consuming. Artificial intelligence (AI) shows promise in detecting prostate cancer and grading patterns. We tested an AI-based digital twin of a pathologist, vPatho, on 2,603 histology images of prostate tissue stained with hematoxylin and eosin. We analyzed various factors influencing tumor-grade disagreement between vPatho and six human pathologists. Our results demonstrated that vPatho achieved comparable performance in prostate cancer detection and tumor volume estimation, as reported in the literature. Concordance levels between vPatho and human pathologists were examined. Notably, moderate to substantial agreement was observed in identifying complementary histological features such as ductal, cribriform, nerve, blood vessels, and lymph cell infiltrations. However, concordance in tumor grading showed a decline when applied to prostatectomy specimens (kappa = 0.44) compared to biopsy cores (kappa = 0.70). Adjusting the decision threshold for the secondary Gleason pattern from 5% to 10% improved the concordance level between pathologists and vPatho for tumor grading on prostatectomy specimens (kappa from 0.44 to 0.64). Potential causes of grade discordance included the vertical extent of tumors toward the prostate boundary and the proportions of slides with prostate cancer. Gleason pattern 4 was particularly associated with discordance. Notably, grade discordance with vPatho was not specific to any of the six pathologists involved in routine clinical grading. In conclusion, our study highlights the potential utility of AI in developing a digital twin of a pathologist. This approach can help uncover limitations in AI adoption and the current grading system for prostate cancer pathology.
Federated Stain Normalization for Computational Pathology
Sep 29, 2022Abstract:Although deep federated learning has received much attention in recent years, progress has been made mainly in the context of natural images and barely for computational pathology. However, deep federated learning is an opportunity to create datasets that reflect the data diversity of many laboratories. Further, the effort of dataset construction can be divided among many. Unfortunately, existing algorithms cannot be easily applied to computational pathology since previous work presupposes that data distributions of laboratories must be similar. This is an unlikely assumption, mainly since different laboratories have different staining styles. As a solution, we propose BottleGAN, a generative model that can computationally align the staining styles of many laboratories and can be trained in a privacy-preserving manner to foster federated learning in computational pathology. We construct a heterogenic multi-institutional dataset based on the PESO segmentation dataset and improve the IOU by 42\% compared to existing federated learning algorithms. An implementation of BottleGAN is available at https://github.com/MECLabTUDA/BottleGAN
Biologic and Prognostic Feature Scores from Whole-Slide Histology Images Using Deep Learning
Oct 24, 2019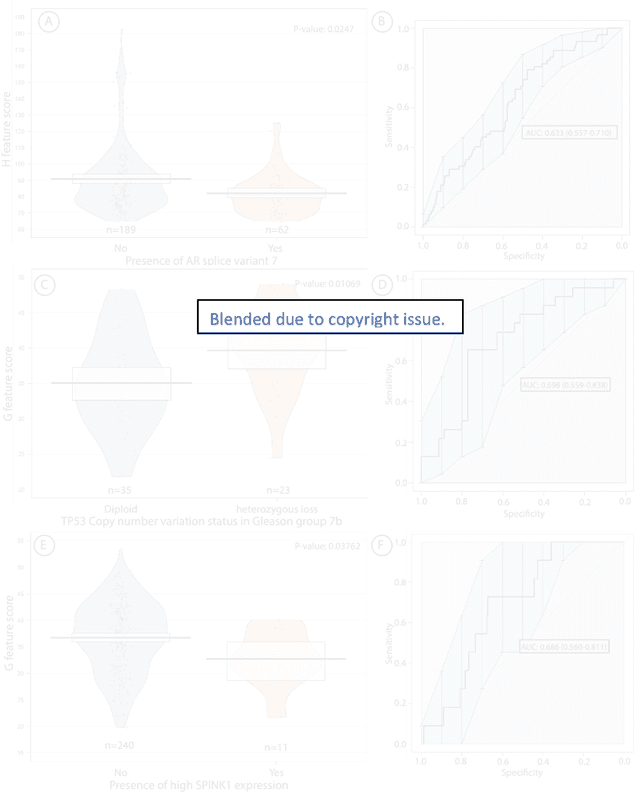
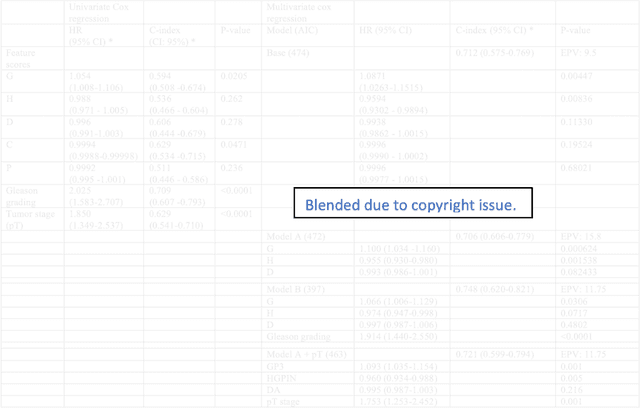
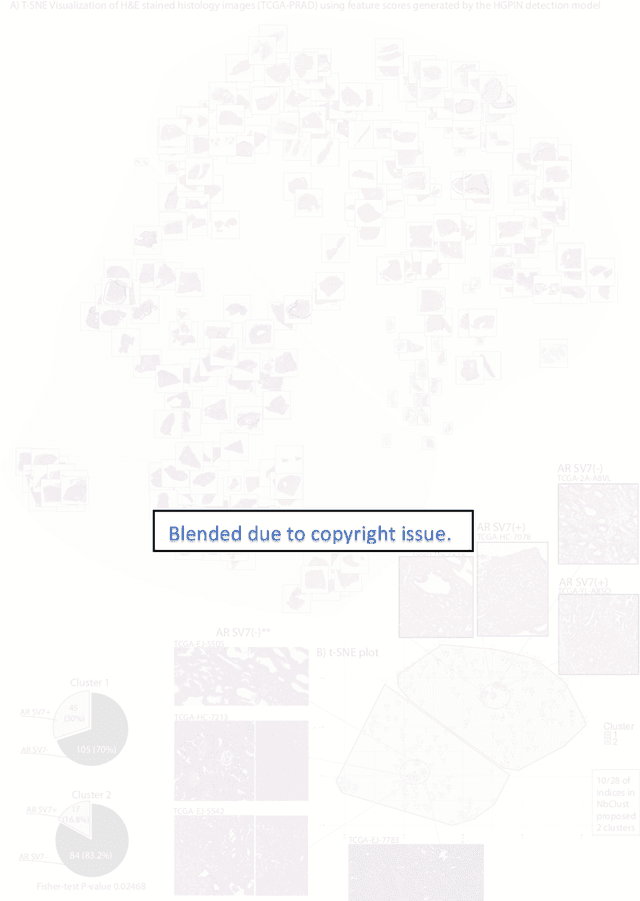
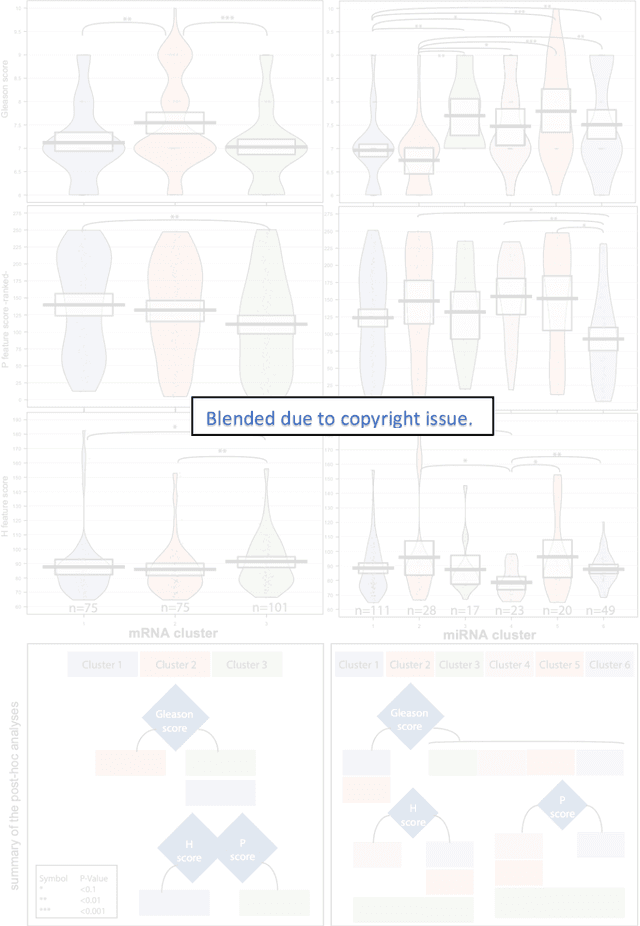
Abstract:Histopathology is a reflection of the molecular changes and provides prognostic phenotypes representing the disease progression. In this study, we introduced feature scores generated from hematoxylin and eosin histology images based on deep learning (DL) models developed for prostate pathology. We demonstrated that these feature scores were significantly prognostic for time to event endpoints (biochemical recurrence and cancer-specific survival) and had simultaneously molecular biologic associations to relevant genomic alterations and molecular subtypes using already trained DL models that were not previously exposed to the datasets of the current study. Further, we discussed the potential of such feature scores to improve the current tumor grading system and the challenges that are associated with tumor heterogeneity and the development of prognostic models from histology images. Our findings uncover the potential of feature scores from histology images as digital biomarkers in precision medicine and as an expanding utility for digital pathology.
Deep Learning for Prostate Pathology
Oct 16, 2019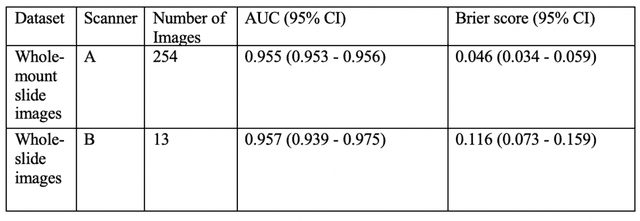
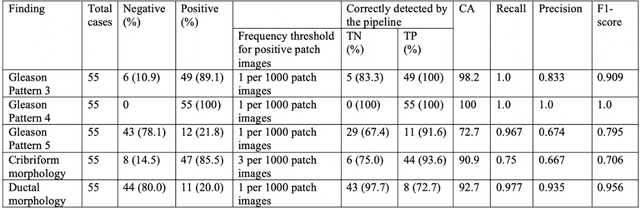
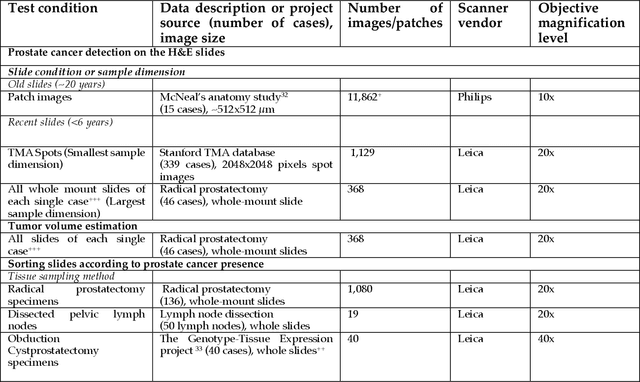
Abstract:The current study detects different morphologies related to prostate pathology using deep learning models; these models were evaluated on 2,121 hematoxylin and eosin (H&E) stain histology images captured using bright field microscopy, which spanned a variety of image qualities, origins (whole slide, tissue micro array, whole mount, Internet), scanning machines, timestamps, H&E staining protocols, and institutions. For case usage, these models were applied for the annotation tasks in clinician-oriented pathology reports for prostatectomy specimens. The true positive rate (TPR) for slides with prostate cancer was 99.7% by a false positive rate of 0.785%. The F1-scores of Gleason patterns reported in pathology reports ranged from 0.795 to 1.0 at the case level. TPR was 93.6% for the cribriform morphology and 72.6% for the ductal morphology. The correlation between the ground truth and the prediction for the relative tumor volume was 0.987 n. Our models cover the major components of prostate pathology and successfully accomplish the annotation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge