Anirban Mukhopadhyay
OctreeNCA: Single-Pass 184 MP Segmentation on Consumer Hardware
Aug 09, 2025Abstract:Medical applications demand segmentation of large inputs, like prostate MRIs, pathology slices, or videos of surgery. These inputs should ideally be inferred at once to provide the model with proper spatial or temporal context. When segmenting large inputs, the VRAM consumption of the GPU becomes the bottleneck. Architectures like UNets or Vision Transformers scale very poorly in VRAM consumption, resulting in patch- or frame-wise approaches that compromise global consistency and inference speed. The lightweight Neural Cellular Automaton (NCA) is a bio-inspired model that is by construction size-invariant. However, due to its local-only communication rules, it lacks global knowledge. We propose OctreeNCA by generalizing the neighborhood definition using an octree data structure. Our generalized neighborhood definition enables the efficient traversal of global knowledge. Since deep learning frameworks are mainly developed for large multi-layer networks, their implementation does not fully leverage the advantages of NCAs. We implement an NCA inference function in CUDA that further reduces VRAM demands and increases inference speed. Our OctreeNCA segments high-resolution images and videos quickly while occupying 90% less VRAM than a UNet during evaluation. This allows us to segment 184 Megapixel pathology slices or 1-minute surgical videos at once.
Don't Reach for the Stars: Rethinking Topology for Resilient Federated Learning
Aug 07, 2025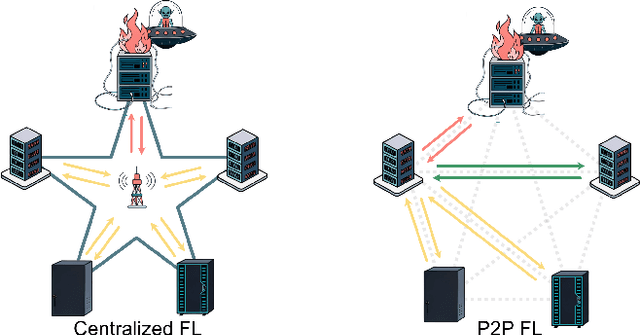
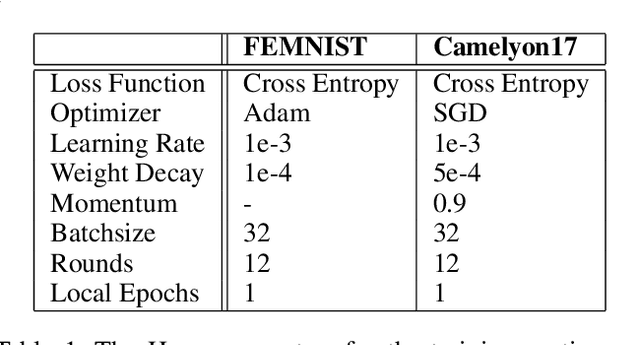
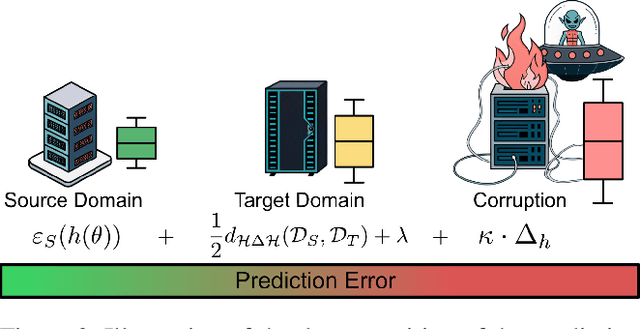
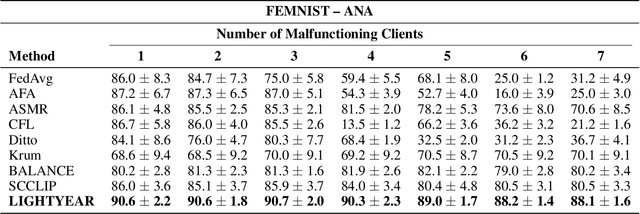
Abstract:Federated learning (FL) enables collaborative model training across distributed clients while preserving data privacy by keeping data local. Traditional FL approaches rely on a centralized, star-shaped topology, where a central server aggregates model updates from clients. However, this architecture introduces several limitations, including a single point of failure, limited personalization, and poor robustness to distribution shifts or vulnerability to malfunctioning clients. Moreover, update selection in centralized FL often relies on low-level parameter differences, which can be unreliable when client data is not independent and identically distributed, and offer clients little control. In this work, we propose a decentralized, peer-to-peer (P2P) FL framework. It leverages the flexibility of the P2P topology to enable each client to identify and aggregate a personalized set of trustworthy and beneficial updates.This framework is the Local Inference Guided Aggregation for Heterogeneous Training Environments to Yield Enhancement Through Agreement and Regularization (LIGHTYEAR). Central to our method is an agreement score, computed on a local validation set, which quantifies the semantic alignment of incoming updates in the function space with respect to the clients reference model. Each client uses this score to select a tailored subset of updates and performs aggregation with a regularization term that further stabilizes the training. Our empirical evaluation across two datasets shows that the proposed approach consistently outperforms both centralized baselines and existing P2P methods in terms of client-level performance, particularly under adversarial and heterogeneous conditions.
eNCApsulate: NCA for Precision Diagnosis on Capsule Endoscopes
Apr 30, 2025
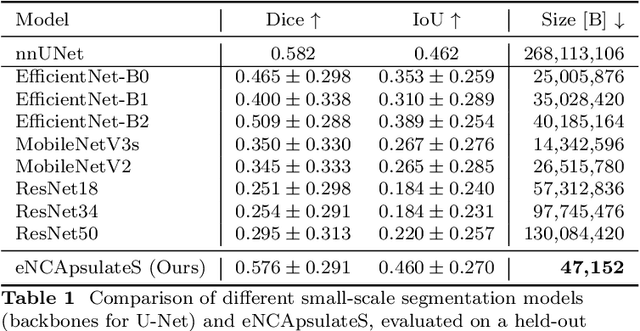
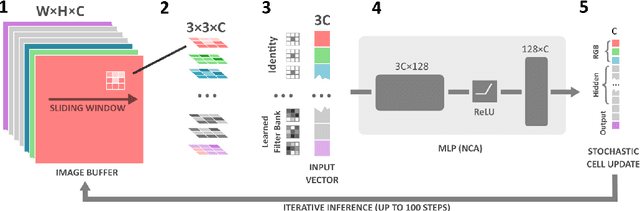
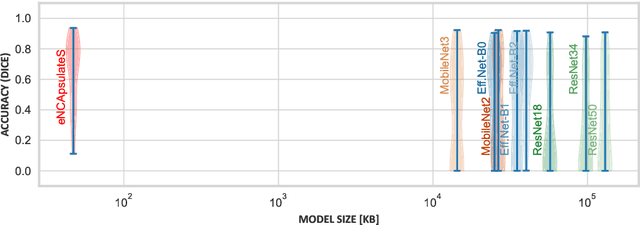
Abstract:Wireless Capsule Endoscopy is a non-invasive imaging method for the entire gastrointestinal tract, and is a pain-free alternative to traditional endoscopy. It generates extensive video data that requires significant review time, and localizing the capsule after ingestion is a challenge. Techniques like bleeding detection and depth estimation can help with localization of pathologies, but deep learning models are typically too large to run directly on the capsule. Neural Cellular Automata (NCA) for bleeding segmentation and depth estimation are trained on capsule endoscopic images. For monocular depth estimation, we distill a large foundation model into the lean NCA architecture, by treating the outputs of the foundation model as pseudo ground truth. We then port the trained NCA to the ESP32 microcontroller, enabling efficient image processing on hardware as small as a camera capsule. NCA are more accurate (Dice) than other portable segmentation models, while requiring more than 100x fewer parameters stored in memory than other small-scale models. The visual results of NCA depth estimation look convincing, and in some cases beat the realism and detail of the pseudo ground truth. Runtime optimizations on the ESP32-S3 accelerate the average inference speed significantly, by more than factor 3. With several algorithmic adjustments and distillation, it is possible to eNCApsulate NCA models into microcontrollers that fit into wireless capsule endoscopes. This is the first work that enables reliable bleeding segmentation and depth estimation on a miniaturized device, paving the way for precise diagnosis combined with visual odometry as a means of precise localization of the capsule -- on the capsule.
SurGrID: Controllable Surgical Simulation via Scene Graph to Image Diffusion
Feb 11, 2025Abstract:Surgical simulation offers a promising addition to conventional surgical training. However, available simulation tools lack photorealism and rely on hardcoded behaviour. Denoising Diffusion Models are a promising alternative for high-fidelity image synthesis, but existing state-of-the-art conditioning methods fall short in providing precise control or interactivity over the generated scenes. We introduce SurGrID, a Scene Graph to Image Diffusion Model, allowing for controllable surgical scene synthesis by leveraging Scene Graphs. These graphs encode a surgical scene's components' spatial and semantic information, which are then translated into an intermediate representation using our novel pre-training step that explicitly captures local and global information. Our proposed method improves the fidelity of generated images and their coherence with the graph input over the state-of-the-art. Further, we demonstrate the simulation's realism and controllability in a user assessment study involving clinical experts. Scene Graphs can be effectively used for precise and interactive conditioning of Denoising Diffusion Models for simulating surgical scenes, enabling high fidelity and interactive control over the generated content.
GAUDA: Generative Adaptive Uncertainty-guided Diffusion-based Augmentation for Surgical Segmentation
Jan 18, 2025



Abstract:Augmentation by generative modelling yields a promising alternative to the accumulation of surgical data, where ethical, organisational and regulatory aspects must be considered. Yet, the joint synthesis of (image, mask) pairs for segmentation, a major application in surgery, is rather unexplored. We propose to learn semantically comprehensive yet compact latent representations of the (image, mask) space, which we jointly model with a Latent Diffusion Model. We show that our approach can effectively synthesise unseen high-quality paired segmentation data of remarkable semantic coherence. Generative augmentation is typically applied pre-training by synthesising a fixed number of additional training samples to improve downstream task models. To enhance this approach, we further propose Generative Adaptive Uncertainty-guided Diffusion-based Augmentation (GAUDA), leveraging the epistemic uncertainty of a Bayesian downstream model for targeted online synthesis. We condition the generative model on classes with high estimated uncertainty during training to produce additional unseen samples for these classes. By adaptively utilising the generative model online, we can minimise the number of additional training samples and centre them around the currently most uncertain parts of the data distribution. GAUDA effectively improves downstream segmentation results over comparable methods by an average absolute IoU of 1.6% on CaDISv2 and 1.5% on CholecSeg8k, two prominent surgical datasets for semantic segmentation.
Federated-Continual Dynamic Segmentation of Histopathology guided by Barlow Continuity
Jan 08, 2025



Abstract:Federated- and Continual Learning have been established as approaches to enable privacy-aware learning on continuously changing data, as required for deploying AI systems in histopathology images. However, data shifts can occur in a dynamic world, spatially between institutions and temporally, due to changing data over time. This leads to two issues: Client Drift, where the central model degrades from aggregating data from clients trained on shifted data, and Catastrophic Forgetting, from temporal shifts such as changes in patient populations. Both tend to degrade the model's performance of previously seen data or spatially distributed training. Despite both problems arising from the same underlying problem of data shifts, existing research addresses them only individually. In this work, we introduce a method that can jointly alleviate Client Drift and Catastrophic Forgetting by using our proposed Dynamic Barlow Continuity that evaluates client updates on a public reference dataset and uses this to guide the training process to a spatially and temporally shift-invariant model. We evaluate our approach on the histopathology datasets BCSS and Semicol and prove our method to be highly effective by jointly improving the dice score as much as from 15.8% to 71.6% in Client Drift and from 42.5% to 62.8% in Catastrophic Forgetting. This enables Dynamic Learning by establishing spatio-temporal shift-invariance.
MedSegDiffNCA: Diffusion Models With Neural Cellular Automata for Skin Lesion Segmentation
Jan 05, 2025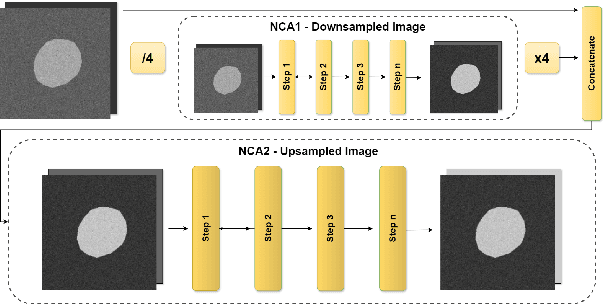
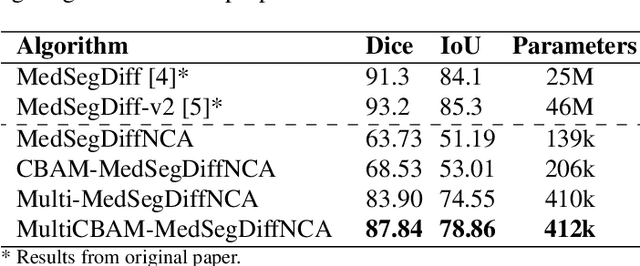
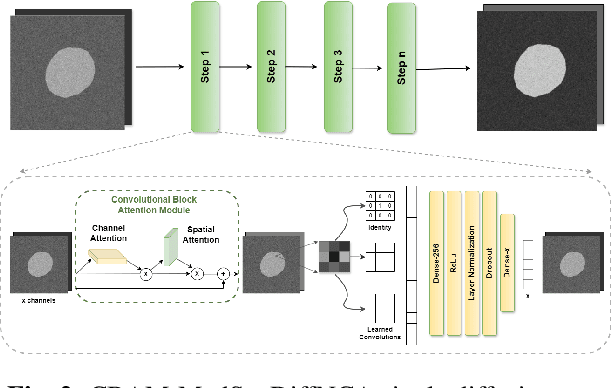

Abstract:Denoising Diffusion Models (DDMs) are widely used for high-quality image generation and medical image segmentation but often rely on Unet-based architectures, leading to high computational overhead, especially with high-resolution images. This work proposes three NCA-based improvements for diffusion-based medical image segmentation. First, Multi-MedSegDiffNCA uses a multilevel NCA framework to refine rough noise estimates generated by lower level NCA models. Second, CBAM-MedSegDiffNCA incorporates channel and spatial attention for improved segmentation. Third, MultiCBAM-MedSegDiffNCA combines these methods with a new RGB channel loss for semantic guidance. Evaluations on Lesion segmentation show that MultiCBAM-MedSegDiffNCA matches Unet-based model performance with dice score of 87.84% while using 60-110 times fewer parameters, offering a more efficient solution for low resource medical settings.
Federated Voxel Scene Graph for Intracranial Hemorrhage
Nov 01, 2024
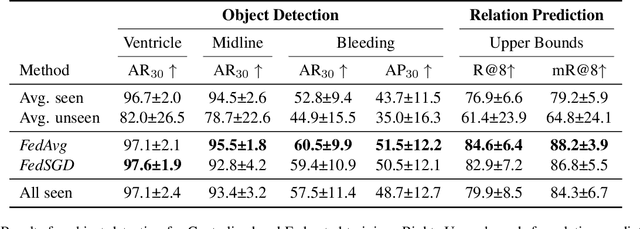
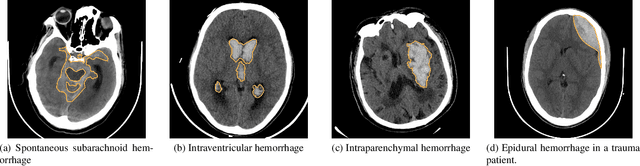
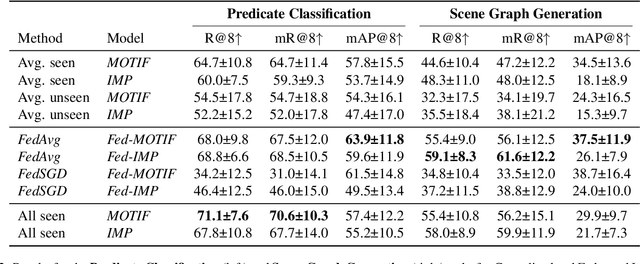
Abstract:Intracranial Hemorrhage is a potentially lethal condition whose manifestation is vastly diverse and shifts across clinical centers worldwide. Deep-learning-based solutions are starting to model complex relations between brain structures, but still struggle to generalize. While gathering more diverse data is the most natural approach, privacy regulations often limit the sharing of medical data. We propose the first application of Federated Scene Graph Generation. We show that our models can leverage the increased training data diversity. For Scene Graph Generation, they can recall up to 20% more clinically relevant relations across datasets compared to models trained on a single centralized dataset. Learning structured data representation in a federated setting can open the way to the development of new methods that can leverage this finer information to regularize across clients more effectively.
NCAdapt: Dynamic adaptation with domain-specific Neural Cellular Automata for continual hippocampus segmentation
Oct 30, 2024Abstract:Continual learning (CL) in medical imaging presents a unique challenge, requiring models to adapt to new domains while retaining previously acquired knowledge. We introduce NCAdapt, a Neural Cellular Automata (NCA) based method designed to address this challenge. NCAdapt features a domain-specific multi-head structure, integrating adaptable convolutional layers into the NCA backbone for each new domain encountered. After initial training, the NCA backbone is frozen, and only the newly added adaptable convolutional layers, consisting of 384 parameters, are trained along with domain-specific NCA convolutions. We evaluate NCAdapt on hippocampus segmentation tasks, benchmarking its performance against Lifelong nnU-Net and U-Net models with state-of-the-art (SOTA) CL methods. Our lightweight approach achieves SOTA performance, underscoring its effectiveness in addressing CL challenges in medical imaging. Upon acceptance, we will make our code base publicly accessible to support reproducibility and foster further advancements in medical CL.
NCA-Morph: Medical Image Registration with Neural Cellular Automata
Oct 29, 2024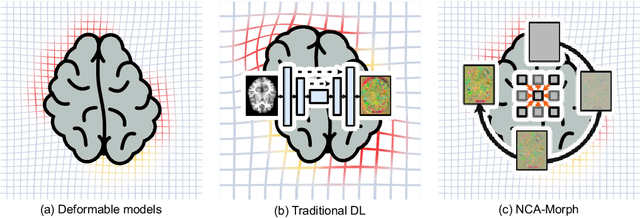
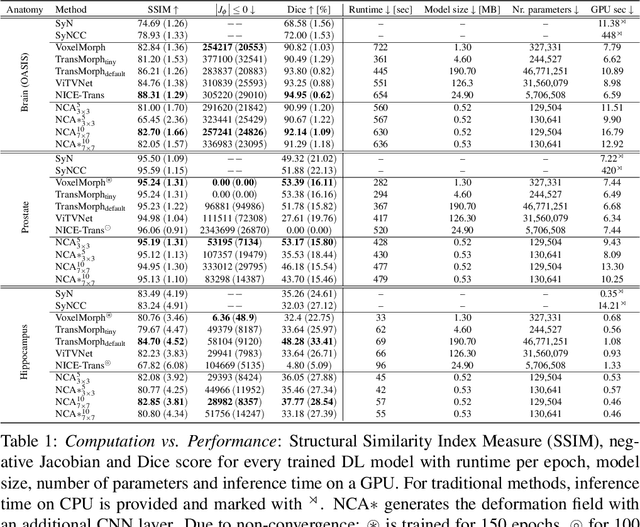
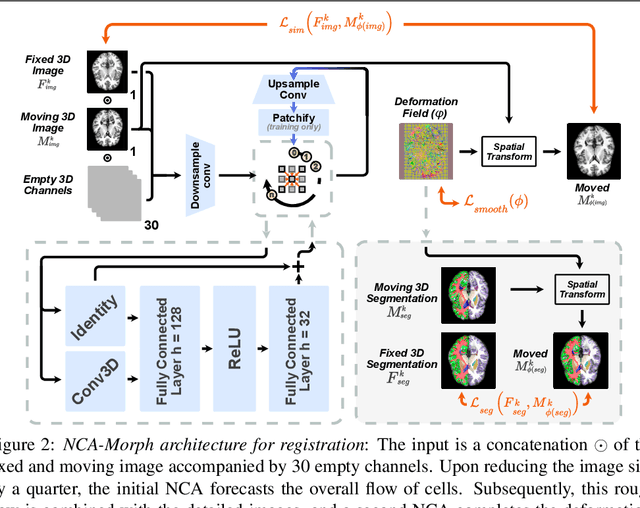
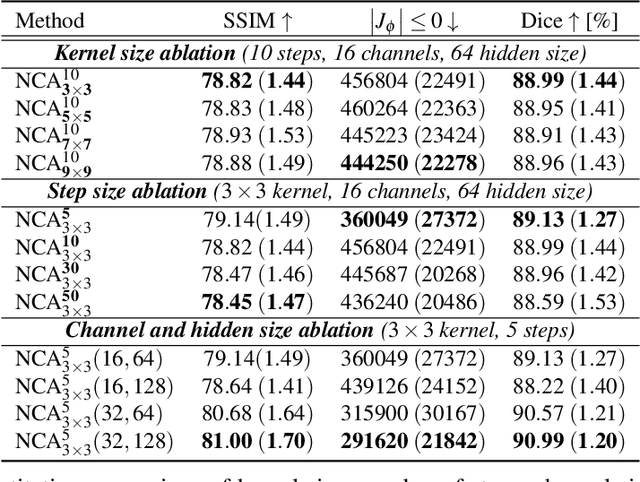
Abstract:Medical image registration is a critical process that aligns various patient scans, facilitating tasks like diagnosis, surgical planning, and tracking. Traditional optimization based methods are slow, prompting the use of Deep Learning (DL) techniques, such as VoxelMorph and Transformer-based strategies, for faster results. However, these DL methods often impose significant resource demands. In response to these challenges, we present NCA-Morph, an innovative approach that seamlessly blends DL with a bio-inspired communication and networking approach, enabled by Neural Cellular Automata (NCAs). NCA-Morph not only harnesses the power of DL for efficient image registration but also builds a network of local communications between cells and respective voxels over time, mimicking the interaction observed in living systems. In our extensive experiments, we subject NCA-Morph to evaluations across three distinct 3D registration tasks, encompassing Brain, Prostate and Hippocampus images from both healthy and diseased patients. The results showcase NCA-Morph's ability to achieve state-of-the-art performance. Notably, NCA-Morph distinguishes itself as a lightweight architecture with significantly fewer parameters; 60% and 99.7% less than VoxelMorph and TransMorph. This characteristic positions NCA-Morph as an ideal solution for resource-constrained medical applications, such as primary care settings and operating rooms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge