Amin Ranem
NCAdapt: Dynamic adaptation with domain-specific Neural Cellular Automata for continual hippocampus segmentation
Oct 30, 2024Abstract:Continual learning (CL) in medical imaging presents a unique challenge, requiring models to adapt to new domains while retaining previously acquired knowledge. We introduce NCAdapt, a Neural Cellular Automata (NCA) based method designed to address this challenge. NCAdapt features a domain-specific multi-head structure, integrating adaptable convolutional layers into the NCA backbone for each new domain encountered. After initial training, the NCA backbone is frozen, and only the newly added adaptable convolutional layers, consisting of 384 parameters, are trained along with domain-specific NCA convolutions. We evaluate NCAdapt on hippocampus segmentation tasks, benchmarking its performance against Lifelong nnU-Net and U-Net models with state-of-the-art (SOTA) CL methods. Our lightweight approach achieves SOTA performance, underscoring its effectiveness in addressing CL challenges in medical imaging. Upon acceptance, we will make our code base publicly accessible to support reproducibility and foster further advancements in medical CL.
NCA-Morph: Medical Image Registration with Neural Cellular Automata
Oct 29, 2024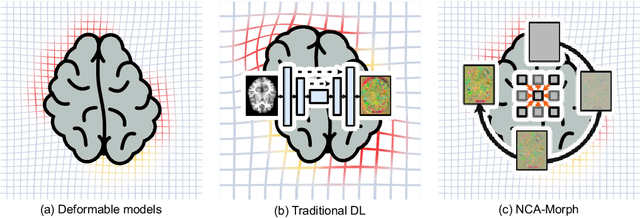
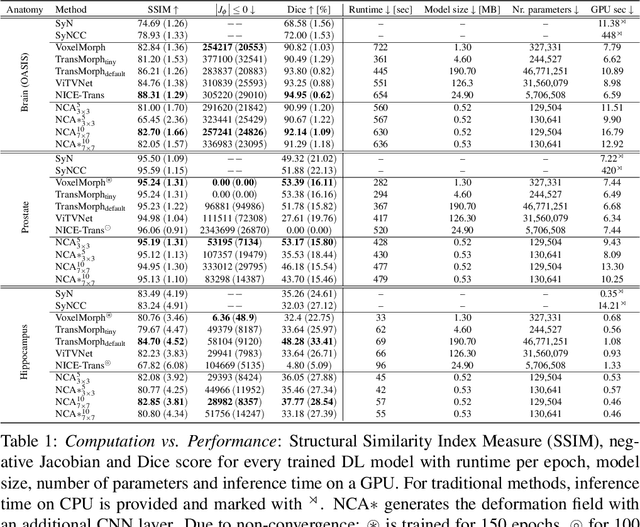
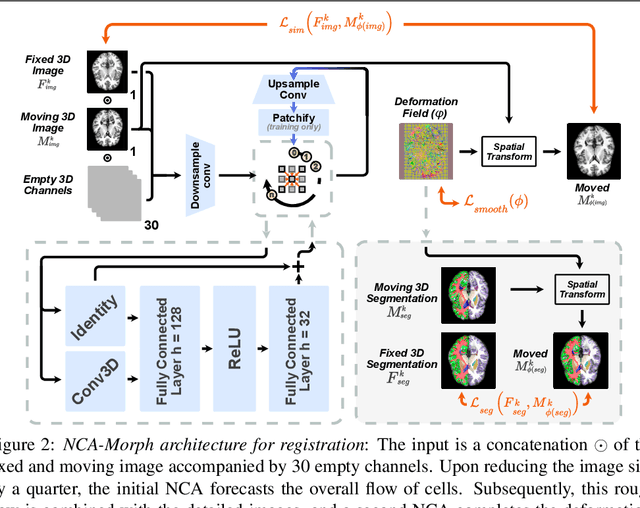
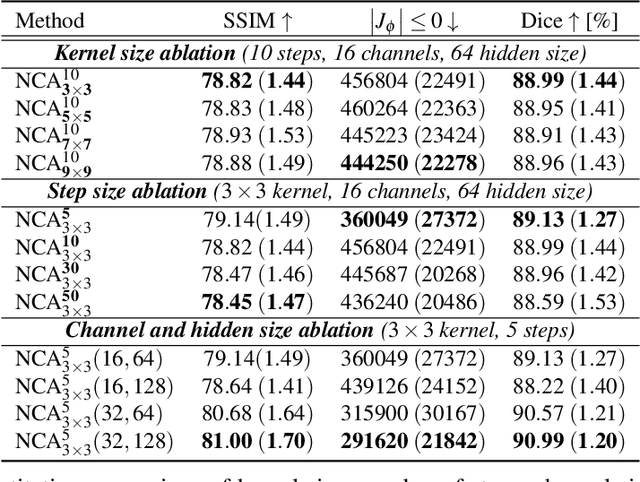
Abstract:Medical image registration is a critical process that aligns various patient scans, facilitating tasks like diagnosis, surgical planning, and tracking. Traditional optimization based methods are slow, prompting the use of Deep Learning (DL) techniques, such as VoxelMorph and Transformer-based strategies, for faster results. However, these DL methods often impose significant resource demands. In response to these challenges, we present NCA-Morph, an innovative approach that seamlessly blends DL with a bio-inspired communication and networking approach, enabled by Neural Cellular Automata (NCAs). NCA-Morph not only harnesses the power of DL for efficient image registration but also builds a network of local communications between cells and respective voxels over time, mimicking the interaction observed in living systems. In our extensive experiments, we subject NCA-Morph to evaluations across three distinct 3D registration tasks, encompassing Brain, Prostate and Hippocampus images from both healthy and diseased patients. The results showcase NCA-Morph's ability to achieve state-of-the-art performance. Notably, NCA-Morph distinguishes itself as a lightweight architecture with significantly fewer parameters; 60% and 99.7% less than VoxelMorph and TransMorph. This characteristic positions NCA-Morph as an ideal solution for resource-constrained medical applications, such as primary care settings and operating rooms.
Unsupervised Training of Neural Cellular Automata on Edge Devices
Jul 25, 2024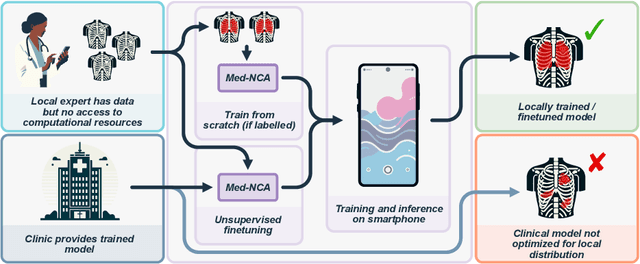
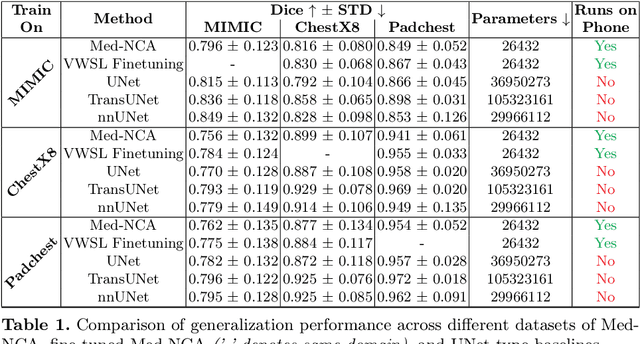
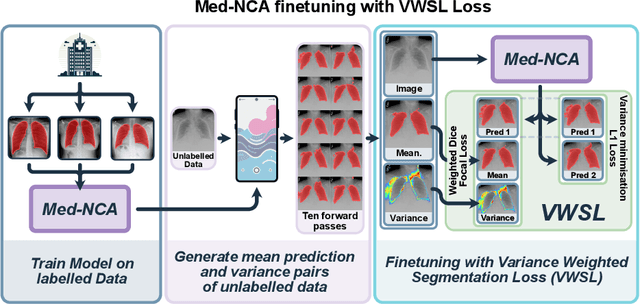
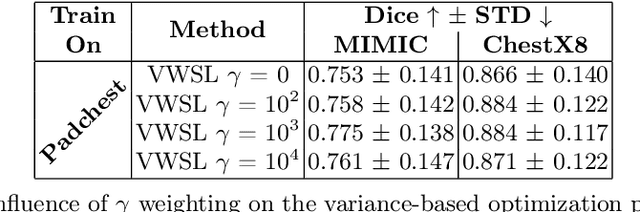
Abstract:The disparity in access to machine learning tools for medical imaging across different regions significantly limits the potential for universal healthcare innovation, particularly in remote areas. Our research addresses this issue by implementing Neural Cellular Automata (NCA) training directly on smartphones for accessible X-ray lung segmentation. We confirm the practicality and feasibility of deploying and training these advanced models on five Android devices, improving medical diagnostics accessibility and bridging the tech divide to extend machine learning benefits in medical imaging to low- and middle-income countries (LMICs). We further enhance this approach with an unsupervised adaptation method using the novel Variance-Weighted Segmentation Loss (VWSL), which efficiently learns from unlabeled data by minimizing the variance from multiple NCA predictions. This strategy notably improves model adaptability and performance across diverse medical imaging contexts without the need for extensive computational resources or labeled datasets, effectively lowering the participation threshold. Our methodology, tested on three multisite X-ray datasets -- Padchest, ChestX-ray8, and MIMIC-III -- demonstrates improvements in segmentation Dice accuracy by 0.7 to 2.8%, compared to the classic Med-NCA. Additionally, in extreme cases where no digital copy is available and images must be captured by a phone from an X-ray lightbox or monitor, VWSL enhances Dice accuracy by 5-20%, demonstrating the method's robustness even with suboptimal image sources.
Continual atlas-based segmentation of prostate MRI
Nov 06, 2023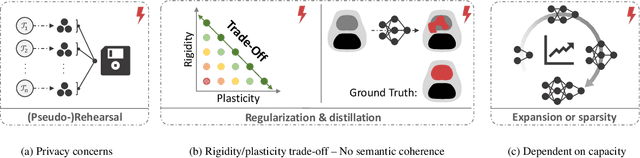
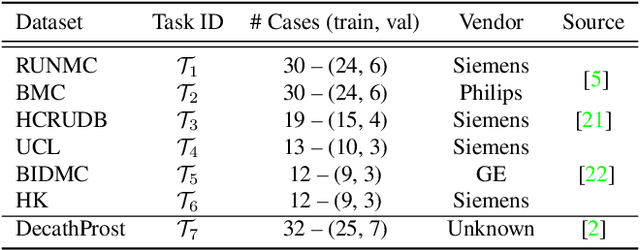
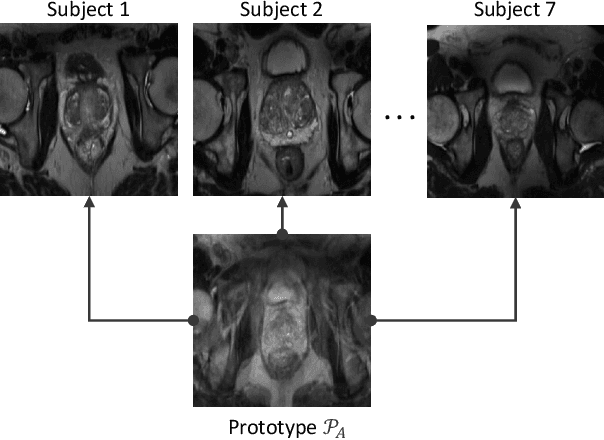
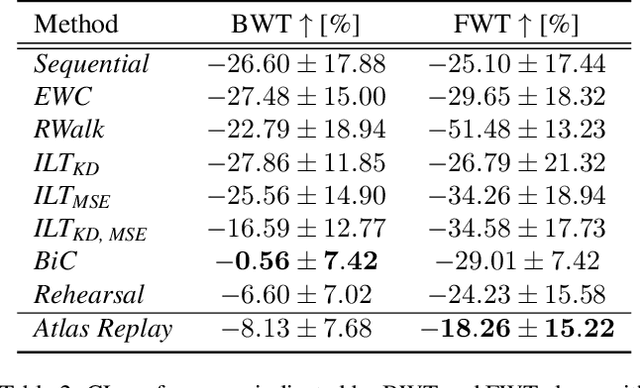
Abstract:Continual learning (CL) methods designed for natural image classification often fail to reach basic quality standards for medical image segmentation. Atlas-based segmentation, a well-established approach in medical imaging, incorporates domain knowledge on the region of interest, leading to semantically coherent predictions. This is especially promising for CL, as it allows us to leverage structural information and strike an optimal balance between model rigidity and plasticity over time. When combined with privacy-preserving prototypes, this process offers the advantages of rehearsal-based CL without compromising patient privacy. We propose Atlas Replay, an atlas-based segmentation approach that uses prototypes to generate high-quality segmentation masks through image registration that maintain consistency even as the training distribution changes. We explore how our proposed method performs compared to state-of-the-art CL methods in terms of knowledge transferability across seven publicly available prostate segmentation datasets. Prostate segmentation plays a vital role in diagnosing prostate cancer, however, it poses challenges due to substantial anatomical variations, benign structural differences in older age groups, and fluctuating acquisition parameters. Our results show that Atlas Replay is both robust and generalizes well to yet-unseen domains while being able to maintain knowledge, unlike end-to-end segmentation methods. Our code base is available under https://github.com/MECLabTUDA/Atlas-Replay.
Exploring SAM Ablations for Enhancing Medical Segmentation in Radiology and Pathology
Sep 30, 2023



Abstract:Medical imaging plays a critical role in the diagnosis and treatment planning of various medical conditions, with radiology and pathology heavily reliant on precise image segmentation. The Segment Anything Model (SAM) has emerged as a promising framework for addressing segmentation challenges across different domains. In this white paper, we delve into SAM, breaking down its fundamental components and uncovering the intricate interactions between them. We also explore the fine-tuning of SAM and assess its profound impact on the accuracy and reliability of segmentation results, focusing on applications in radiology (specifically, brain tumor segmentation) and pathology (specifically, breast cancer segmentation). Through a series of carefully designed experiments, we analyze SAM's potential application in the field of medical imaging. We aim to bridge the gap between advanced segmentation techniques and the demanding requirements of healthcare, shedding light on SAM's transformative capabilities.
Detecting respiratory motion artefacts for cardiovascular MRIs to ensure high-quality segmentation
Sep 20, 2022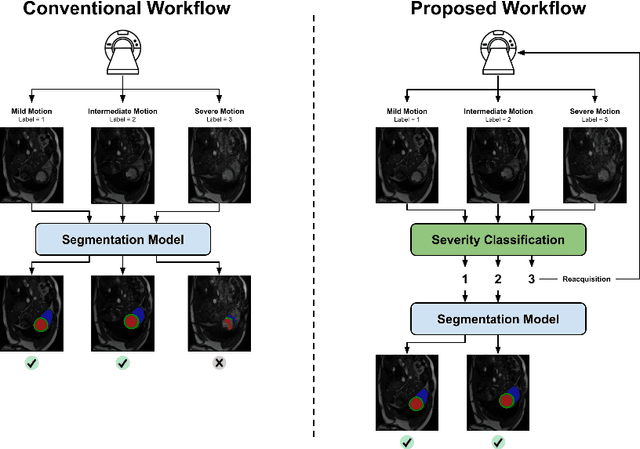
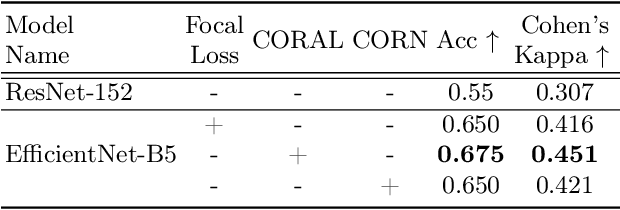
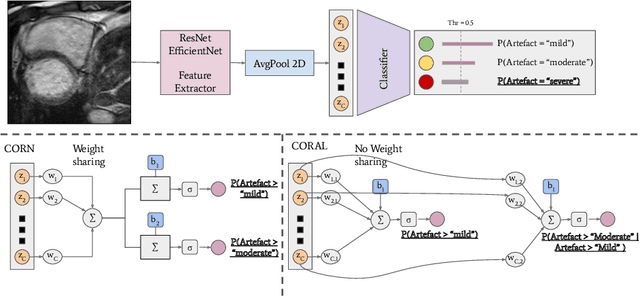
Abstract:While machine learning approaches perform well on their training domain, they generally tend to fail in a real-world application. In cardiovascular magnetic resonance imaging (CMR), respiratory motion represents a major challenge in terms of acquisition quality and therefore subsequent analysis and final diagnosis. We present a workflow which predicts a severity score for respiratory motion in CMR for the CMRxMotion challenge 2022. This is an important tool for technicians to immediately provide feedback on the CMR quality during acquisition, as poor-quality images can directly be re-acquired while the patient is still available in the vicinity. Thus, our method ensures that the acquired CMR holds up to a specific quality standard before it is used for further diagnosis. Therefore, it enables an efficient base for proper diagnosis without having time and cost-intensive re-acquisitions in cases of severe motion artefacts. Combined with our segmentation model, this can help cardiologists and technicians in their daily routine by providing a complete pipeline to guarantee proper quality assessment and genuine segmentations for cardiovascular scans. The code base is available at https://github.com/MECLabTUDA/QA_med_data/tree/dev_QA_CMRxMotion.
Task-agnostic Continual Hippocampus Segmentation for Smooth Population Shifts
Aug 05, 2022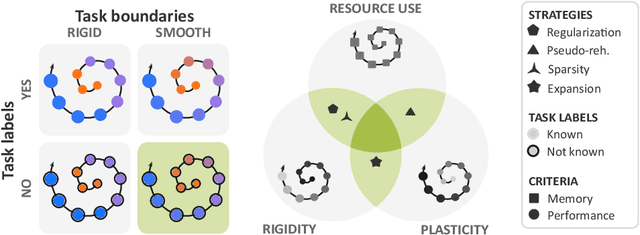
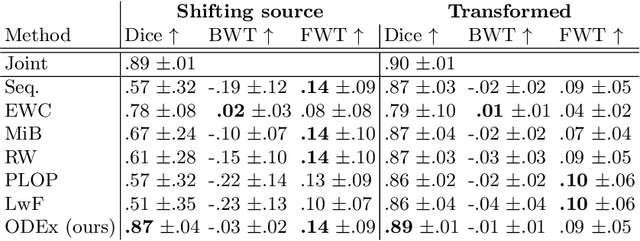
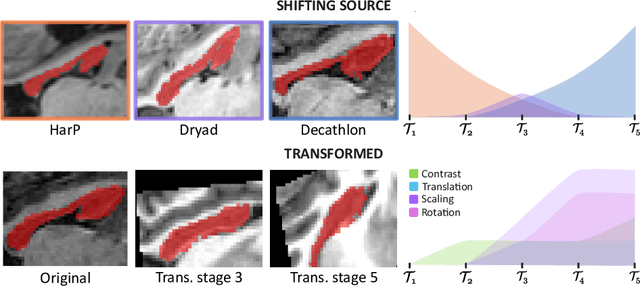
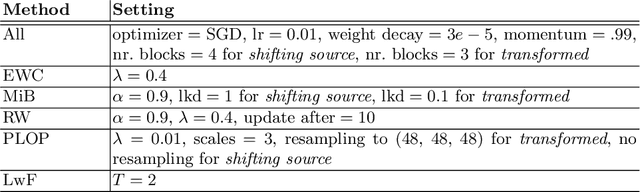
Abstract:Most continual learning methods are validated in settings where task boundaries are clearly defined and task identity information is available during training and testing. We explore how such methods perform in a task-agnostic setting that more closely resembles dynamic clinical environments with gradual population shifts. We propose ODEx, a holistic solution that combines out-of-distribution detection with continual learning techniques. Validation on two scenarios of hippocampus segmentation shows that our proposed method reliably maintains performance on earlier tasks without losing plasticity.
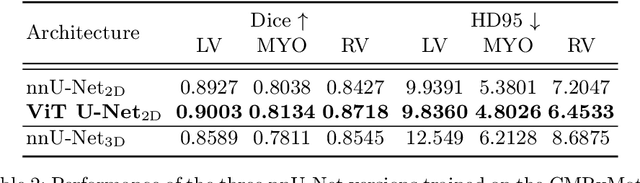
Continual Hippocampus Segmentation with Transformers
Apr 17, 2022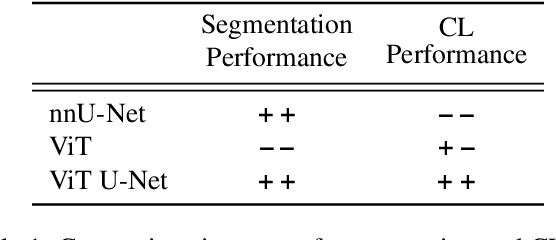
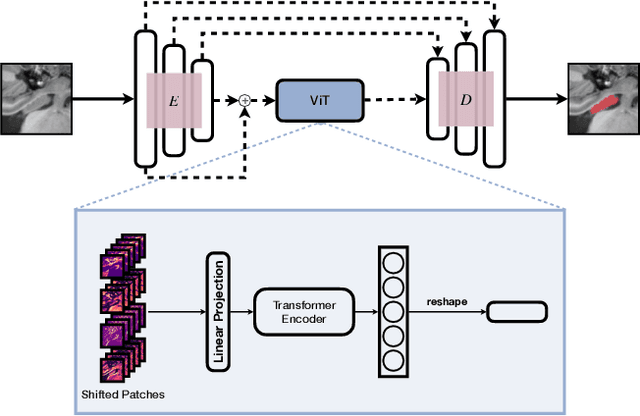
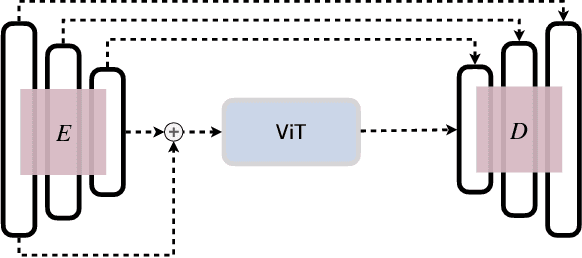
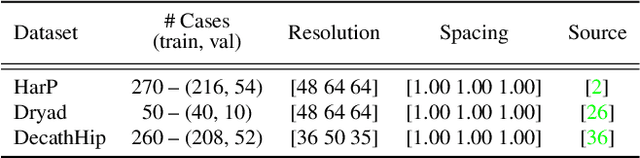
Abstract:In clinical settings, where acquisition conditions and patient populations change over time, continual learning is key for ensuring the safe use of deep neural networks. Yet most existing work focuses on convolutional architectures and image classification. Instead, radiologists prefer to work with segmentation models that outline specific regions-of-interest, for which Transformer-based architectures are gaining traction. The self-attention mechanism of Transformers could potentially mitigate catastrophic forgetting, opening the way for more robust medical image segmentation. In this work, we explore how recently-proposed Transformer mechanisms for semantic segmentation behave in sequential learning scenarios, and analyse how best to adapt continual learning strategies for this setting. Our evaluation on hippocampus segmentation shows that Transformer mechanisms mitigate catastrophic forgetting for medical image segmentation compared to purely convolutional architectures, and demonstrates that regularising ViT modules should be done with caution.
Quality monitoring of federated Covid-19 lesion segmentation
Dec 16, 2021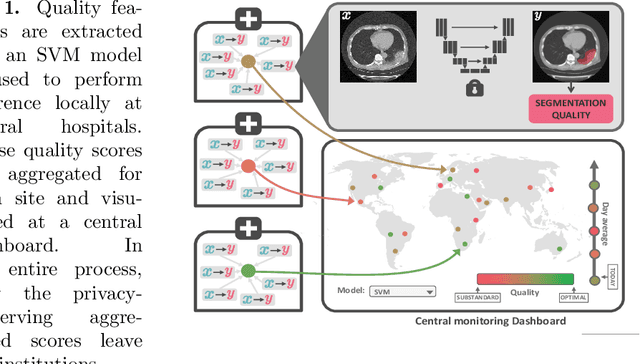
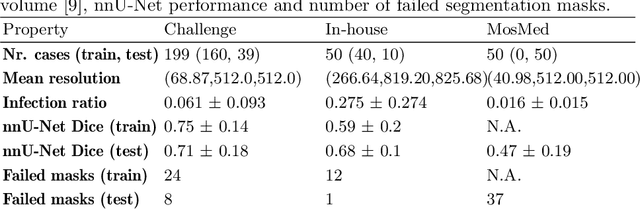
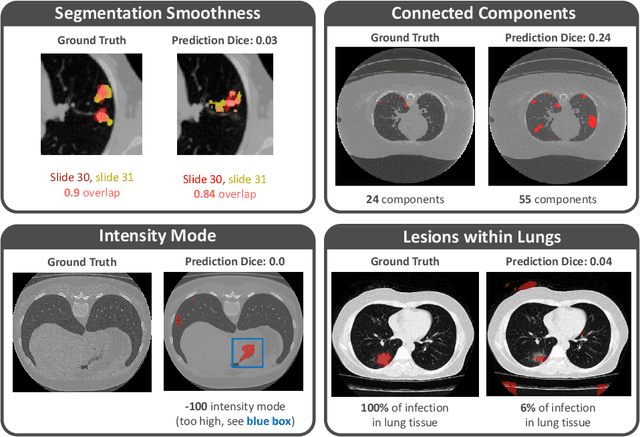
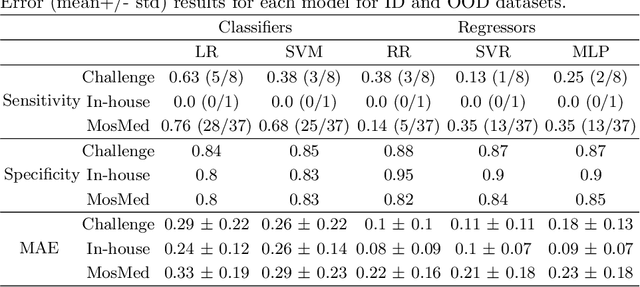
Abstract:Federated Learning is the most promising way to train robust Deep Learning models for the segmentation of Covid-19-related findings in chest CTs. By learning in a decentralized fashion, heterogeneous data can be leveraged from a variety of sources and acquisition protocols whilst ensuring patient privacy. It is, however, crucial to continuously monitor the performance of the model. Yet when it comes to the segmentation of diffuse lung lesions, a quick visual inspection is not enough to assess the quality, and thorough monitoring of all network outputs by expert radiologists is not feasible. In this work, we present an array of lightweight metrics that can be calculated locally in each hospital and then aggregated for central monitoring of a federated system. Our linear model detects over 70% of low-quality segmentations on an out-of-distribution dataset and thus reliably signals a decline in model performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge