Camila González
In the Picture: Medical Imaging Datasets, Artifacts, and their Living Review
Jan 18, 2025


Abstract:Datasets play a critical role in medical imaging research, yet issues such as label quality, shortcuts, and metadata are often overlooked. This lack of attention may harm the generalizability of algorithms and, consequently, negatively impact patient outcomes. While existing medical imaging literature reviews mostly focus on machine learning (ML) methods, with only a few focusing on datasets for specific applications, these reviews remain static -- they are published once and not updated thereafter. This fails to account for emerging evidence, such as biases, shortcuts, and additional annotations that other researchers may contribute after the dataset is published. We refer to these newly discovered findings of datasets as research artifacts. To address this gap, we propose a living review that continuously tracks public datasets and their associated research artifacts across multiple medical imaging applications. Our approach includes a framework for the living review to monitor data documentation artifacts, and an SQL database to visualize the citation relationships between research artifact and dataset. Lastly, we discuss key considerations for creating medical imaging datasets, review best practices for data annotation, discuss the significance of shortcuts and demographic diversity, and emphasize the importance of managing datasets throughout their entire lifecycle. Our demo is publicly available at http://130.226.140.142.
Distribution-Aware Replay for Continual MRI Segmentation
Jul 30, 2024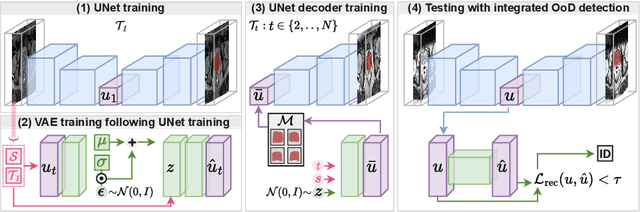
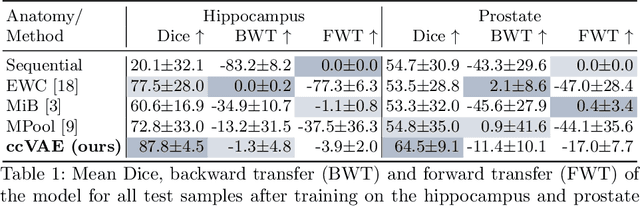
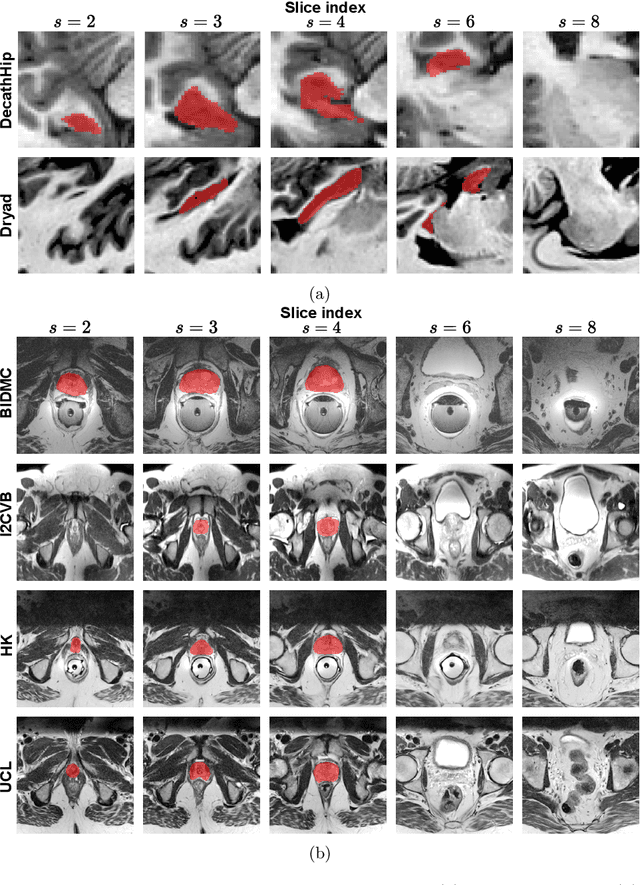
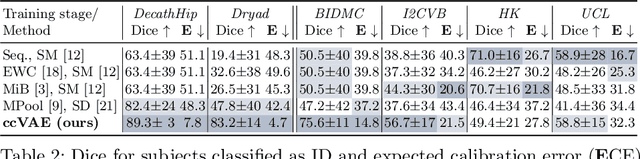
Abstract:Medical image distributions shift constantly due to changes in patient population and discrepancies in image acquisition. These distribution changes result in performance deterioration; deterioration that continual learning aims to alleviate. However, only adaptation with data rehearsal strategies yields practically desirable performance for medical image segmentation. Such rehearsal violates patient privacy and, as most continual learning approaches, overlooks unexpected changes from out-of-distribution instances. To transcend both of these challenges, we introduce a distribution-aware replay strategy that mitigates forgetting through auto-encoding of features, while simultaneously leveraging the learned distribution of features to detect model failure. We provide empirical corroboration on hippocampus and prostate MRI segmentation.
Continual atlas-based segmentation of prostate MRI
Nov 06, 2023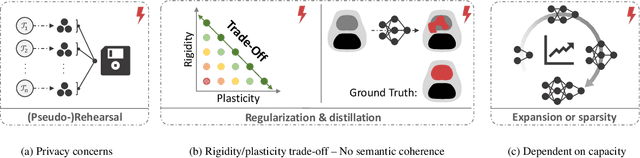
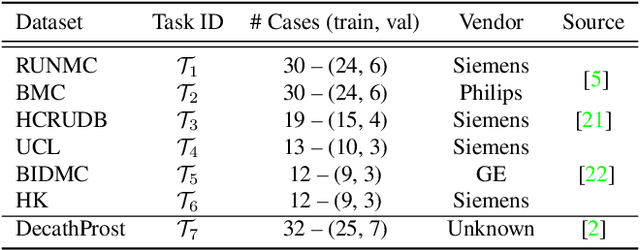
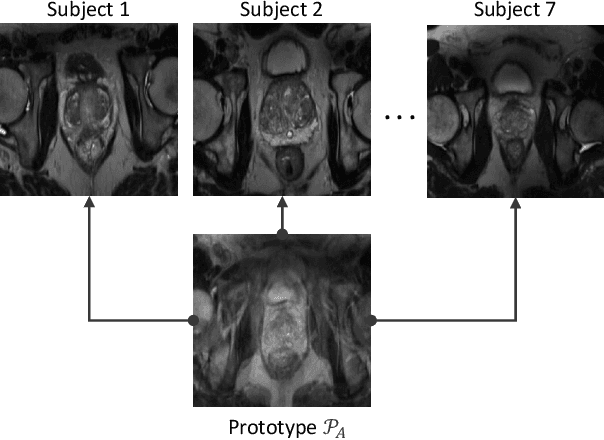
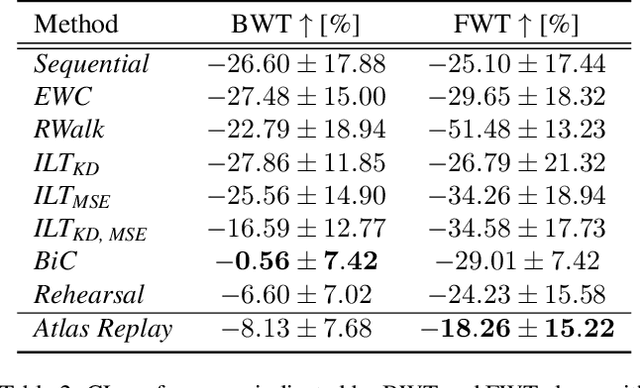
Abstract:Continual learning (CL) methods designed for natural image classification often fail to reach basic quality standards for medical image segmentation. Atlas-based segmentation, a well-established approach in medical imaging, incorporates domain knowledge on the region of interest, leading to semantically coherent predictions. This is especially promising for CL, as it allows us to leverage structural information and strike an optimal balance between model rigidity and plasticity over time. When combined with privacy-preserving prototypes, this process offers the advantages of rehearsal-based CL without compromising patient privacy. We propose Atlas Replay, an atlas-based segmentation approach that uses prototypes to generate high-quality segmentation masks through image registration that maintain consistency even as the training distribution changes. We explore how our proposed method performs compared to state-of-the-art CL methods in terms of knowledge transferability across seven publicly available prostate segmentation datasets. Prostate segmentation plays a vital role in diagnosing prostate cancer, however, it poses challenges due to substantial anatomical variations, benign structural differences in older age groups, and fluctuating acquisition parameters. Our results show that Atlas Replay is both robust and generalizes well to yet-unseen domains while being able to maintain knowledge, unlike end-to-end segmentation methods. Our code base is available under https://github.com/MECLabTUDA/Atlas-Replay.
Med-NCA: Robust and Lightweight Segmentation with Neural Cellular Automata
Feb 07, 2023Abstract:Access to the proper infrastructure is critical when performing medical image segmentation with Deep Learning. This requirement makes it difficult to run state-of-the-art segmentation models in resource-constrained scenarios like primary care facilities in rural areas and during crises. The recently emerging field of Neural Cellular Automata (NCA) has shown that locally interacting one-cell models can achieve competitive results in tasks such as image generation or segmentations in low-resolution inputs. However, they are constrained by high VRAM requirements and the difficulty of reaching convergence for high-resolution images. To counteract these limitations we propose Med-NCA, an end-to-end NCA training pipeline for high-resolution image segmentation. Our method follows a two-step process. Global knowledge is first communicated between cells across the downscaled image. Following that, patch-based segmentation is performed. Our proposed Med-NCA outperforms the classic UNet by 2% and 3% Dice for hippocampus and prostate segmentation, respectively, while also being 500 times smaller. We also show that Med-NCA is by design invariant with respect to image scale, shape and translation, experiencing only slight performance degradation even with strong shifts; and is robust against MRI acquisition artefacts. Med-NCA enables high-resolution medical image segmentation even on a Raspberry Pi B+, arguably the smallest device able to run PyTorch and that can be powered by a standard power bank.
Continual Hippocampus Segmentation with Transformers
Apr 17, 2022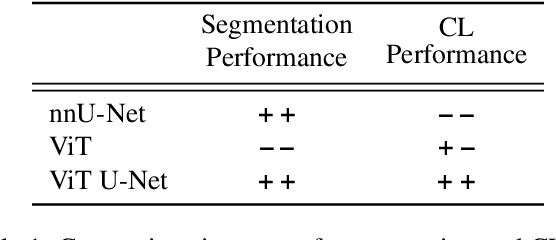
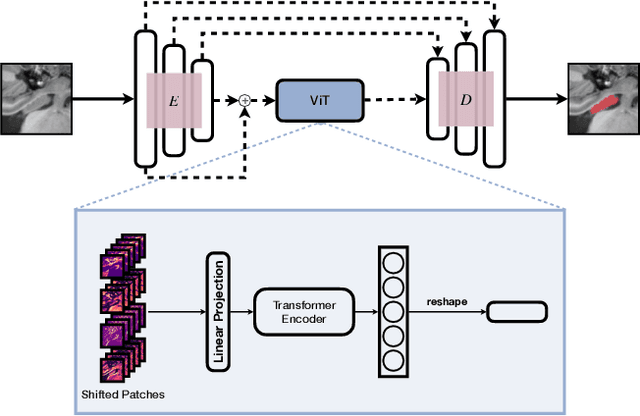
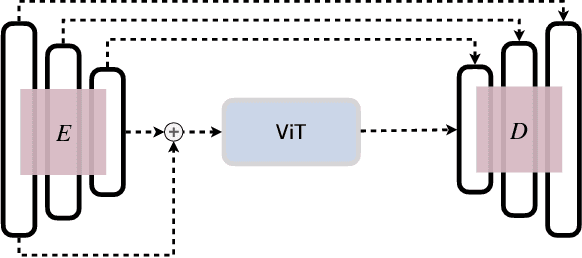
Abstract:In clinical settings, where acquisition conditions and patient populations change over time, continual learning is key for ensuring the safe use of deep neural networks. Yet most existing work focuses on convolutional architectures and image classification. Instead, radiologists prefer to work with segmentation models that outline specific regions-of-interest, for which Transformer-based architectures are gaining traction. The self-attention mechanism of Transformers could potentially mitigate catastrophic forgetting, opening the way for more robust medical image segmentation. In this work, we explore how recently-proposed Transformer mechanisms for semantic segmentation behave in sequential learning scenarios, and analyse how best to adapt continual learning strategies for this setting. Our evaluation on hippocampus segmentation shows that Transformer mechanisms mitigate catastrophic forgetting for medical image segmentation compared to purely convolutional architectures, and demonstrates that regularising ViT modules should be done with caution.
Improving robustness and calibration in ensembles with diversity regularization
Jan 26, 2022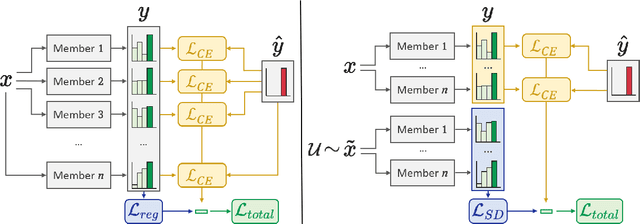

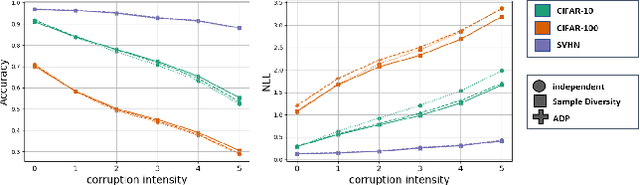

Abstract:Calibration and uncertainty estimation are crucial topics in high-risk environments. We introduce a new diversity regularizer for classification tasks that uses out-of-distribution samples and increases the overall accuracy, calibration and out-of-distribution detection capabilities of ensembles. Following the recent interest in the diversity of ensembles, we systematically evaluate the viability of explicitly regularizing ensemble diversity to improve calibration on in-distribution data as well as under dataset shift. We demonstrate that diversity regularization is highly beneficial in architectures, where weights are partially shared between the individual members and even allows to use fewer ensemble members to reach the same level of robustness. Experiments on CIFAR-10, CIFAR-100, and SVHN show that regularizing diversity can have a significant impact on calibration and robustness, as well as out-of-distribution detection.
Disentanglement enables cross-domain Hippocampus Segmentation
Jan 14, 2022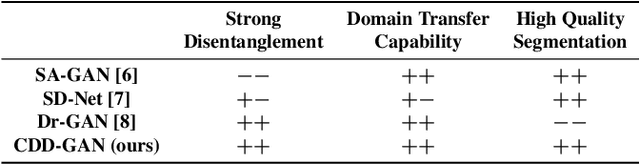
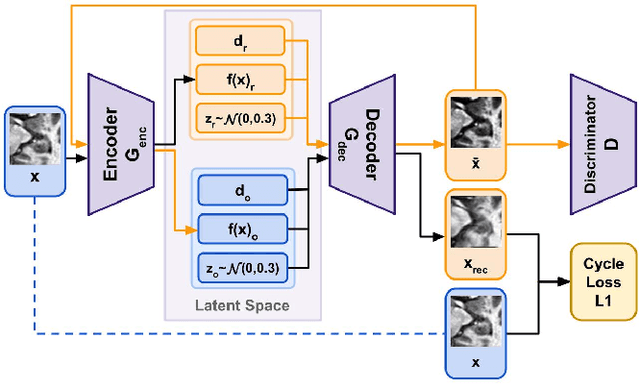
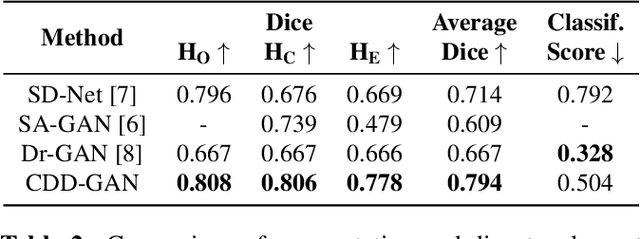
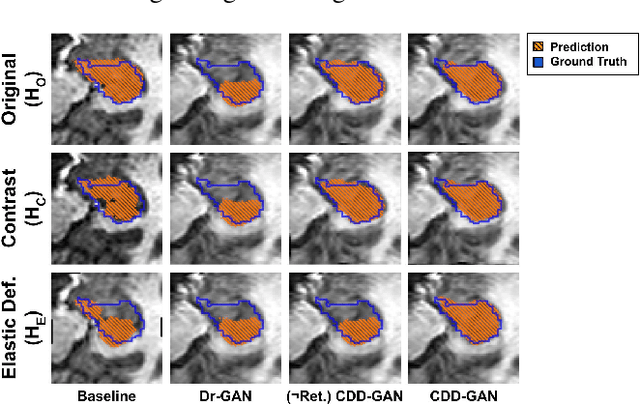
Abstract:Limited amount of labelled training data are a common problem in medical imaging. This makes it difficult to train a well-generalised model and therefore often leads to failure in unknown domains. Hippocampus segmentation from magnetic resonance imaging (MRI) scans is critical for the diagnosis and treatment of neuropsychatric disorders. Domain differences in contrast or shape can significantly affect segmentation. We address this issue by disentangling a T1-weighted MRI image into its content and domain. This separation enables us to perform a domain transfer and thus convert data from new sources into the training domain. This step thus simplifies the segmentation problem, resulting in higher quality segmentations. We achieve the disentanglement with the proposed novel methodology 'Content Domain Disentanglement GAN', and we propose to retrain the UNet on the transformed outputs to deal with GAN-specific artefacts. With these changes, we are able to improve performance on unseen domains by 6-13% and outperform state-of-the-art domain transfer methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge