John Kalkhof
OctreeNCA: Single-Pass 184 MP Segmentation on Consumer Hardware
Aug 09, 2025Abstract:Medical applications demand segmentation of large inputs, like prostate MRIs, pathology slices, or videos of surgery. These inputs should ideally be inferred at once to provide the model with proper spatial or temporal context. When segmenting large inputs, the VRAM consumption of the GPU becomes the bottleneck. Architectures like UNets or Vision Transformers scale very poorly in VRAM consumption, resulting in patch- or frame-wise approaches that compromise global consistency and inference speed. The lightweight Neural Cellular Automaton (NCA) is a bio-inspired model that is by construction size-invariant. However, due to its local-only communication rules, it lacks global knowledge. We propose OctreeNCA by generalizing the neighborhood definition using an octree data structure. Our generalized neighborhood definition enables the efficient traversal of global knowledge. Since deep learning frameworks are mainly developed for large multi-layer networks, their implementation does not fully leverage the advantages of NCAs. We implement an NCA inference function in CUDA that further reduces VRAM demands and increases inference speed. Our OctreeNCA segments high-resolution images and videos quickly while occupying 90% less VRAM than a UNet during evaluation. This allows us to segment 184 Megapixel pathology slices or 1-minute surgical videos at once.
MedSegDiffNCA: Diffusion Models With Neural Cellular Automata for Skin Lesion Segmentation
Jan 05, 2025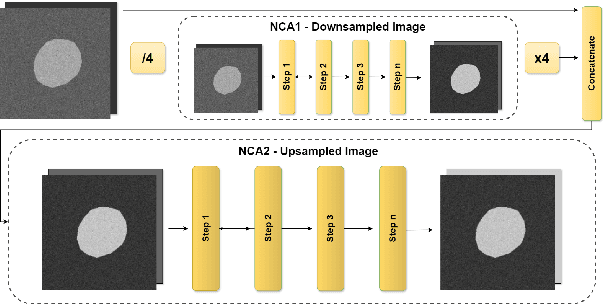
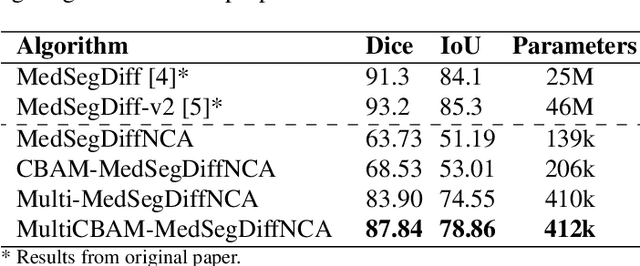
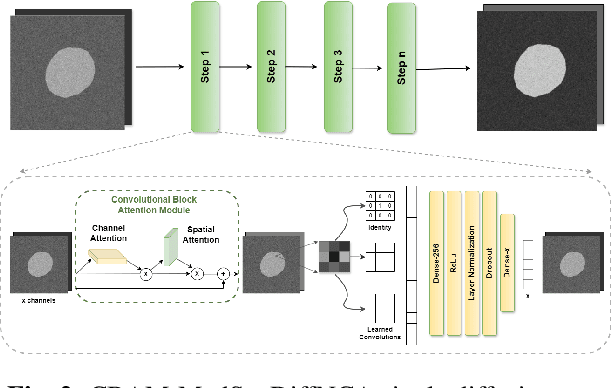

Abstract:Denoising Diffusion Models (DDMs) are widely used for high-quality image generation and medical image segmentation but often rely on Unet-based architectures, leading to high computational overhead, especially with high-resolution images. This work proposes three NCA-based improvements for diffusion-based medical image segmentation. First, Multi-MedSegDiffNCA uses a multilevel NCA framework to refine rough noise estimates generated by lower level NCA models. Second, CBAM-MedSegDiffNCA incorporates channel and spatial attention for improved segmentation. Third, MultiCBAM-MedSegDiffNCA combines these methods with a new RGB channel loss for semantic guidance. Evaluations on Lesion segmentation show that MultiCBAM-MedSegDiffNCA matches Unet-based model performance with dice score of 87.84% while using 60-110 times fewer parameters, offering a more efficient solution for low resource medical settings.
NCAdapt: Dynamic adaptation with domain-specific Neural Cellular Automata for continual hippocampus segmentation
Oct 30, 2024Abstract:Continual learning (CL) in medical imaging presents a unique challenge, requiring models to adapt to new domains while retaining previously acquired knowledge. We introduce NCAdapt, a Neural Cellular Automata (NCA) based method designed to address this challenge. NCAdapt features a domain-specific multi-head structure, integrating adaptable convolutional layers into the NCA backbone for each new domain encountered. After initial training, the NCA backbone is frozen, and only the newly added adaptable convolutional layers, consisting of 384 parameters, are trained along with domain-specific NCA convolutions. We evaluate NCAdapt on hippocampus segmentation tasks, benchmarking its performance against Lifelong nnU-Net and U-Net models with state-of-the-art (SOTA) CL methods. Our lightweight approach achieves SOTA performance, underscoring its effectiveness in addressing CL challenges in medical imaging. Upon acceptance, we will make our code base publicly accessible to support reproducibility and foster further advancements in medical CL.
NCA-Morph: Medical Image Registration with Neural Cellular Automata
Oct 29, 2024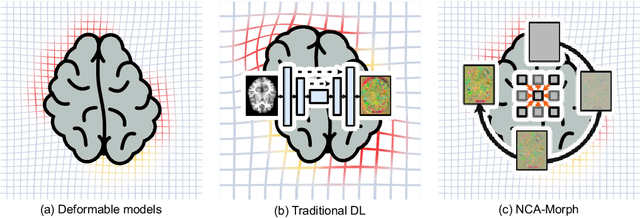
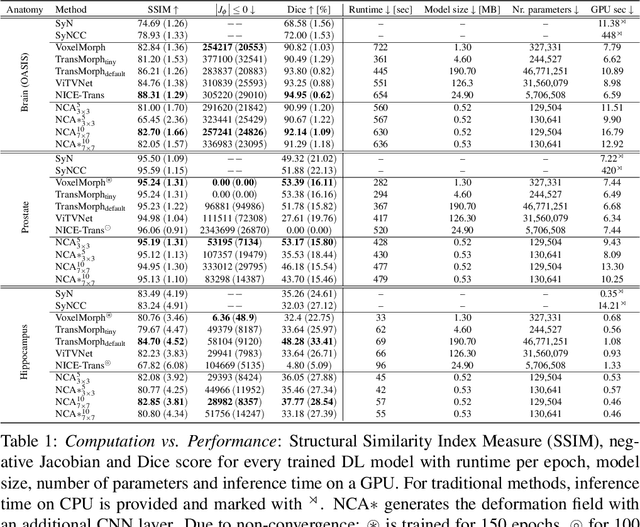
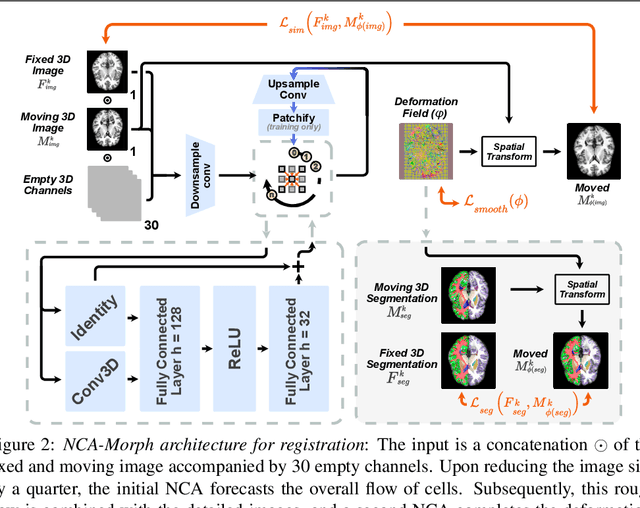
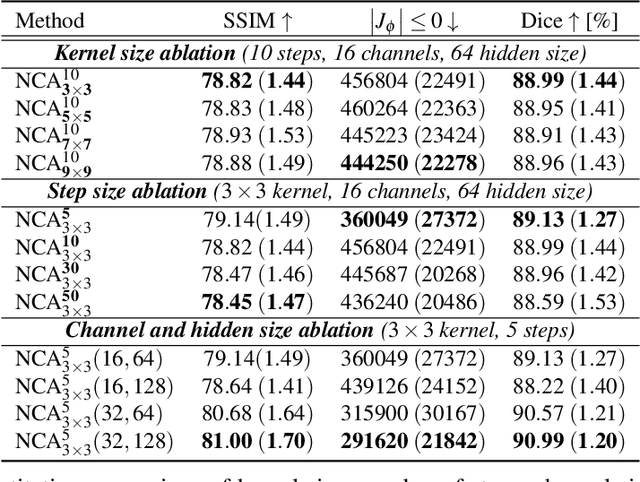
Abstract:Medical image registration is a critical process that aligns various patient scans, facilitating tasks like diagnosis, surgical planning, and tracking. Traditional optimization based methods are slow, prompting the use of Deep Learning (DL) techniques, such as VoxelMorph and Transformer-based strategies, for faster results. However, these DL methods often impose significant resource demands. In response to these challenges, we present NCA-Morph, an innovative approach that seamlessly blends DL with a bio-inspired communication and networking approach, enabled by Neural Cellular Automata (NCAs). NCA-Morph not only harnesses the power of DL for efficient image registration but also builds a network of local communications between cells and respective voxels over time, mimicking the interaction observed in living systems. In our extensive experiments, we subject NCA-Morph to evaluations across three distinct 3D registration tasks, encompassing Brain, Prostate and Hippocampus images from both healthy and diseased patients. The results showcase NCA-Morph's ability to achieve state-of-the-art performance. Notably, NCA-Morph distinguishes itself as a lightweight architecture with significantly fewer parameters; 60% and 99.7% less than VoxelMorph and TransMorph. This characteristic positions NCA-Morph as an ideal solution for resource-constrained medical applications, such as primary care settings and operating rooms.
Unsupervised Training of Neural Cellular Automata on Edge Devices
Jul 25, 2024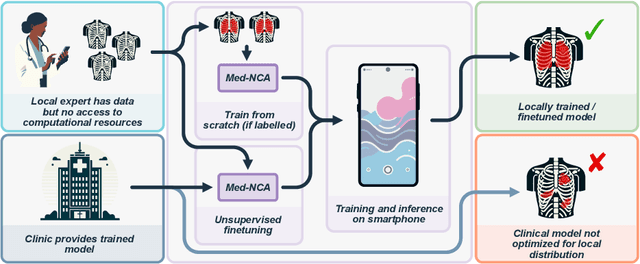
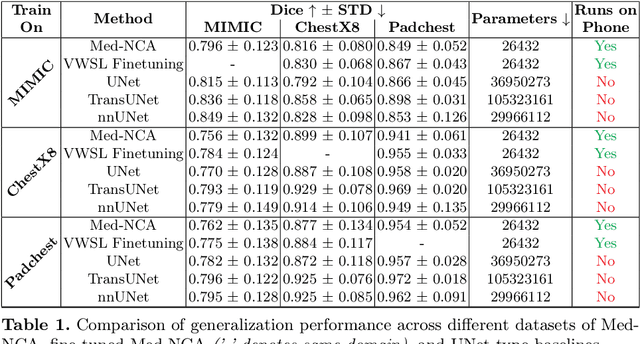
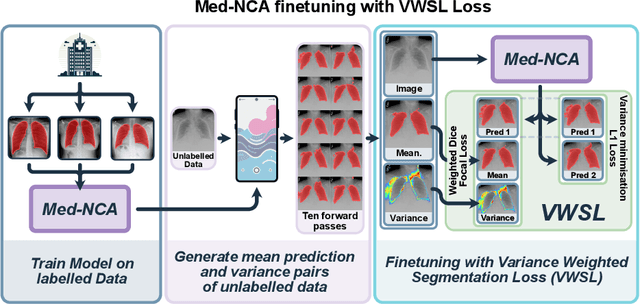
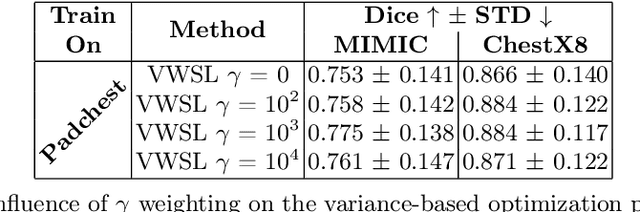
Abstract:The disparity in access to machine learning tools for medical imaging across different regions significantly limits the potential for universal healthcare innovation, particularly in remote areas. Our research addresses this issue by implementing Neural Cellular Automata (NCA) training directly on smartphones for accessible X-ray lung segmentation. We confirm the practicality and feasibility of deploying and training these advanced models on five Android devices, improving medical diagnostics accessibility and bridging the tech divide to extend machine learning benefits in medical imaging to low- and middle-income countries (LMICs). We further enhance this approach with an unsupervised adaptation method using the novel Variance-Weighted Segmentation Loss (VWSL), which efficiently learns from unlabeled data by minimizing the variance from multiple NCA predictions. This strategy notably improves model adaptability and performance across diverse medical imaging contexts without the need for extensive computational resources or labeled datasets, effectively lowering the participation threshold. Our methodology, tested on three multisite X-ray datasets -- Padchest, ChestX-ray8, and MIMIC-III -- demonstrates improvements in segmentation Dice accuracy by 0.7 to 2.8%, compared to the classic Med-NCA. Additionally, in extreme cases where no digital copy is available and images must be captured by a phone from an X-ray lightbox or monitor, VWSL enhances Dice accuracy by 5-20%, demonstrating the method's robustness even with suboptimal image sources.
Frequency-Time Diffusion with Neural Cellular Automata
Jan 11, 2024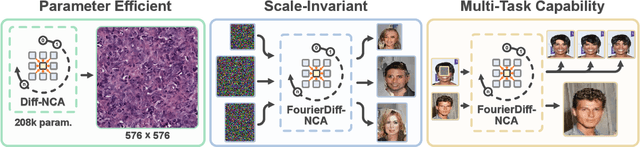
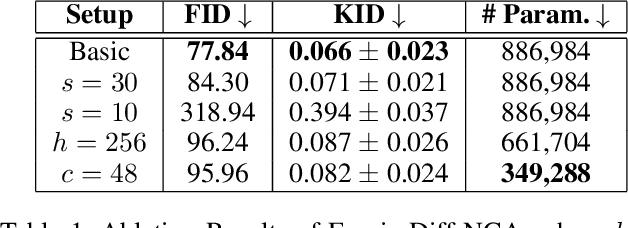
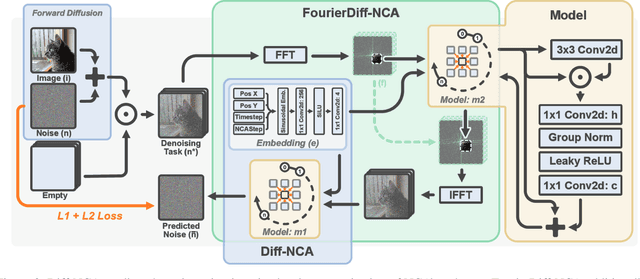
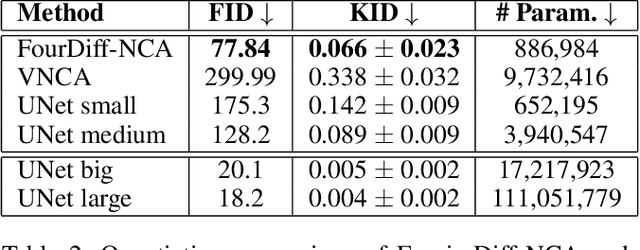
Abstract:Denoising Diffusion Models (DDMs) have become the leading generative technique for synthesizing high-quality images but are often constrained by their UNet-based architectures that impose certain limitations. In particular, the considerable size of often hundreds of millions of parameters makes them impractical when hardware resources are limited. However, even with powerful hardware, processing images in the gigapixel range is difficult. This is especially true in fields such as microscopy or satellite imaging, where such challenges arise from the limitation to a predefined generative size and the inefficient scaling to larger images. We present two variations of Neural Cellular Automata (NCA)-based DDM methods to address these challenges and jumpstart NCA-based DDMs: Diff-NCA and FourierDiff-NCA. Diff-NCA performs diffusion by using only local features of the underlying distribution, making it suitable for applications where local features are critical. To communicate global knowledge in image space, naive NCA setups require timesteps that increase with the image scale. We solve this bottleneck of current NCA architectures by introducing FourierDiff-NCA, which advances Diff-NCA by adding a Fourier-based diffusion process and combines the frequency-organized Fourier space with the image space. By initiating diffusion in the Fourier domain and finalizing it in the image space, FourierDiff-NCA accelerates global communication. We validate our techniques by using Diff-NCA (208k parameters) to generate high-resolution digital pathology scans at 576x576 resolution and FourierDiff-NCA (887k parameters) to synthesize CelebA images at 64x64, outperforming VNCA and five times bigger UNet-based DDMs. In addition, we demonstrate FourierDiff-NCA's capabilities in super-resolution, OOD image synthesis, and inpainting without additional training.
M3D-NCA: Robust 3D Segmentation with Built-in Quality Control
Sep 06, 2023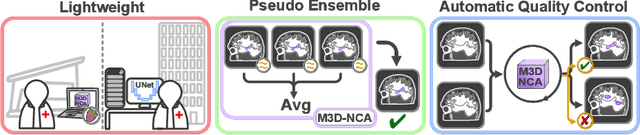
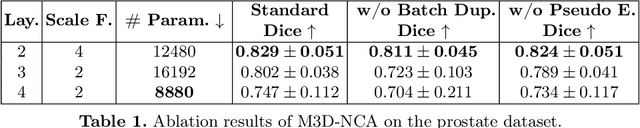


Abstract:Medical image segmentation relies heavily on large-scale deep learning models, such as UNet-based architectures. However, the real-world utility of such models is limited by their high computational requirements, which makes them impractical for resource-constrained environments such as primary care facilities and conflict zones. Furthermore, shifts in the imaging domain can render these models ineffective and even compromise patient safety if such errors go undetected. To address these challenges, we propose M3D-NCA, a novel methodology that leverages Neural Cellular Automata (NCA) segmentation for 3D medical images using n-level patchification. Moreover, we exploit the variance in M3D-NCA to develop a novel quality metric which can automatically detect errors in the segmentation process of NCAs. M3D-NCA outperforms the two magnitudes larger UNet models in hippocampus and prostate segmentation by 2% Dice and can be run on a Raspberry Pi 4 Model B (2GB RAM). This highlights the potential of M3D-NCA as an effective and efficient alternative for medical image segmentation in resource-constrained environments.
Med-NCA: Robust and Lightweight Segmentation with Neural Cellular Automata
Feb 07, 2023Abstract:Access to the proper infrastructure is critical when performing medical image segmentation with Deep Learning. This requirement makes it difficult to run state-of-the-art segmentation models in resource-constrained scenarios like primary care facilities in rural areas and during crises. The recently emerging field of Neural Cellular Automata (NCA) has shown that locally interacting one-cell models can achieve competitive results in tasks such as image generation or segmentations in low-resolution inputs. However, they are constrained by high VRAM requirements and the difficulty of reaching convergence for high-resolution images. To counteract these limitations we propose Med-NCA, an end-to-end NCA training pipeline for high-resolution image segmentation. Our method follows a two-step process. Global knowledge is first communicated between cells across the downscaled image. Following that, patch-based segmentation is performed. Our proposed Med-NCA outperforms the classic UNet by 2% and 3% Dice for hippocampus and prostate segmentation, respectively, while also being 500 times smaller. We also show that Med-NCA is by design invariant with respect to image scale, shape and translation, experiencing only slight performance degradation even with strong shifts; and is robust against MRI acquisition artefacts. Med-NCA enables high-resolution medical image segmentation even on a Raspberry Pi B+, arguably the smallest device able to run PyTorch and that can be powered by a standard power bank.
Detecting respiratory motion artefacts for cardiovascular MRIs to ensure high-quality segmentation
Sep 20, 2022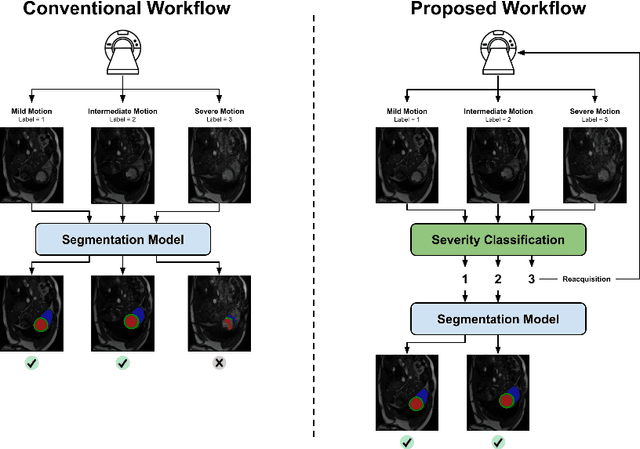
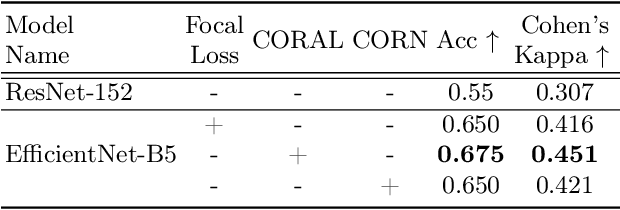
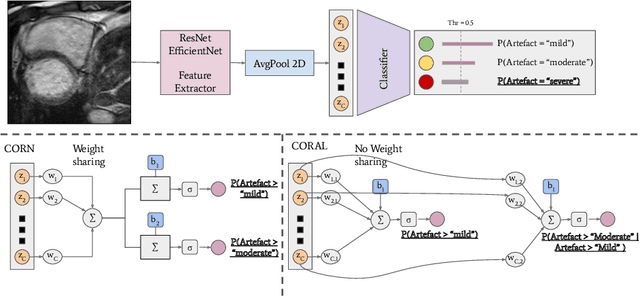
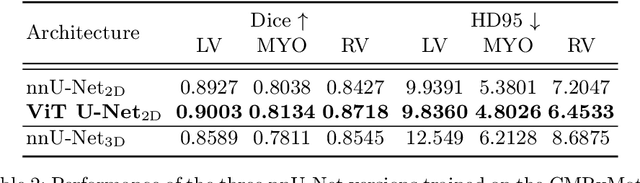
Abstract:While machine learning approaches perform well on their training domain, they generally tend to fail in a real-world application. In cardiovascular magnetic resonance imaging (CMR), respiratory motion represents a major challenge in terms of acquisition quality and therefore subsequent analysis and final diagnosis. We present a workflow which predicts a severity score for respiratory motion in CMR for the CMRxMotion challenge 2022. This is an important tool for technicians to immediately provide feedback on the CMR quality during acquisition, as poor-quality images can directly be re-acquired while the patient is still available in the vicinity. Thus, our method ensures that the acquired CMR holds up to a specific quality standard before it is used for further diagnosis. Therefore, it enables an efficient base for proper diagnosis without having time and cost-intensive re-acquisitions in cases of severe motion artefacts. Combined with our segmentation model, this can help cardiologists and technicians in their daily routine by providing a complete pipeline to guarantee proper quality assessment and genuine segmentations for cardiovascular scans. The code base is available at https://github.com/MECLabTUDA/QA_med_data/tree/dev_QA_CMRxMotion.
Disentanglement enables cross-domain Hippocampus Segmentation
Jan 14, 2022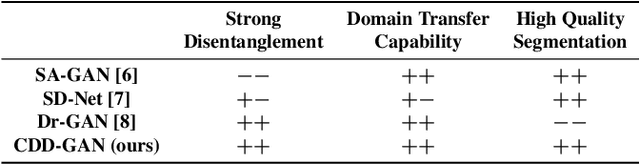
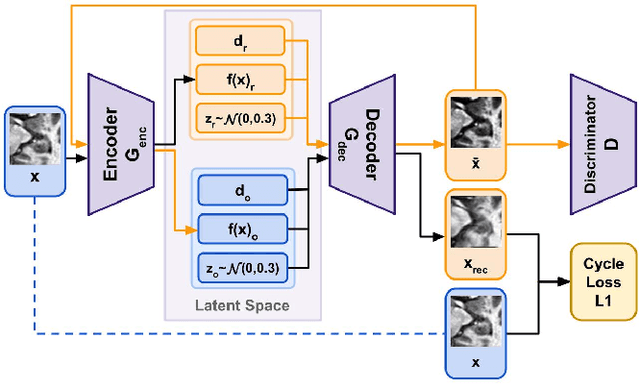
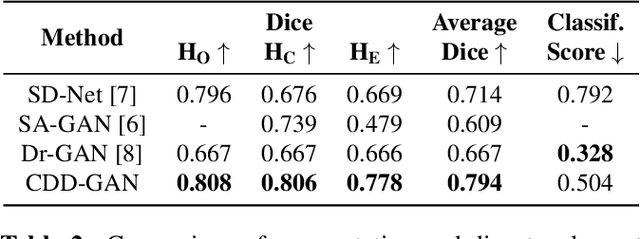
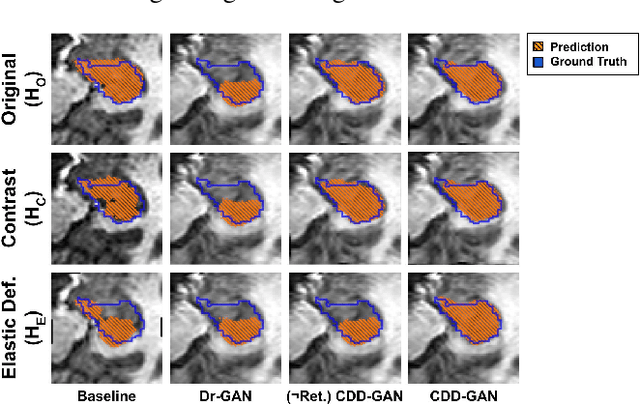
Abstract:Limited amount of labelled training data are a common problem in medical imaging. This makes it difficult to train a well-generalised model and therefore often leads to failure in unknown domains. Hippocampus segmentation from magnetic resonance imaging (MRI) scans is critical for the diagnosis and treatment of neuropsychatric disorders. Domain differences in contrast or shape can significantly affect segmentation. We address this issue by disentangling a T1-weighted MRI image into its content and domain. This separation enables us to perform a domain transfer and thus convert data from new sources into the training domain. This step thus simplifies the segmentation problem, resulting in higher quality segmentations. We achieve the disentanglement with the proposed novel methodology 'Content Domain Disentanglement GAN', and we propose to retrain the UNet on the transformed outputs to deal with GAN-specific artefacts. With these changes, we are able to improve performance on unseen domains by 6-13% and outperform state-of-the-art domain transfer methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge