Arnav Bhavsar
TrackletGPT: A Language-like GPT Framework for White Matter Tract Segmentation
Jan 20, 2026Abstract:White Matter Tract Segmentation is imperative for studying brain structural connectivity, neurological disorders and neurosurgery. This task remains complex, as tracts differ among themselves, across subjects and conditions, yet have similar 3D structure across hemispheres and subjects. To address these challenges, we propose TrackletGPT, a language-like GPT framework which reintroduces sequential information in tokens using tracklets. TrackletGPT generalises seamlessly across datasets, is fully automatic, and encodes granular sub-streamline segments, Tracklets, scaling and refining GPT models in Tractography Segmentation. Based on our experiments, TrackletGPT outperforms state-of-the-art methods on average DICE, Overlap and Overreach scores on TractoInferno and HCP datasets, even on inter-dataset experiments.
TractRLFusion: A GPT-Based Multi-Critic Policy Fusion Framework for Fiber Tractography
Jan 20, 2026Abstract:Tractography plays a pivotal role in the non-invasive reconstruction of white matter fiber pathways, providing vital information on brain connectivity and supporting precise neurosurgical planning. Although traditional methods relied mainly on classical deterministic and probabilistic approaches, recent progress has benefited from supervised deep learning (DL) and deep reinforcement learning (DRL) to improve tract reconstruction. A persistent challenge in tractography is accurately reconstructing white matter tracts while minimizing spurious connections. To address this, we propose TractRLFusion, a novel GPT-based policy fusion framework that integrates multiple RL policies through a data-driven fusion strategy. Our method employs a two-stage training data selection process for effective policy fusion, followed by a multi-critic fine-tuning phase to enhance robustness and generalization. Experiments on HCP, ISMRM, and TractoInferno datasets demonstrate that TractRLFusion outperforms individual RL policies as well as state-of-the-art classical and DRL methods in accuracy and anatomical reliability.
YOLOBirDrone: Dataset for Bird vs Drone Detection and Classification and a YOLO based enhanced learning architecture
Jan 13, 2026Abstract:The use of aerial drones for commercial and defense applications has benefited in many ways and is therefore utilized in several different application domains. However, they are also increasingly used for targeted attacks, posing a significant safety challenge and necessitating the development of drone detection systems. Vision-based drone detection systems currently have an accuracy limitation and struggle to distinguish between drones and birds, particularly when the birds are small in size. This research work proposes a novel YOLOBirDrone architecture that improves the detection and classification accuracy of birds and drones. YOLOBirDrone has different components, including an adaptive and extended layer aggregation (AELAN), a multi-scale progressive dual attention module (MPDA), and a reverse MPDA (RMPDA) to preserve shape information and enrich features with local and global spatial and channel information. A large-scale dataset, BirDrone, is also introduced in this article, which includes small and challenging objects for robust aerial object identification. Experimental results demonstrate an improvement in performance metrics through the proposed YOLOBirDrone architecture compared to other state-of-the-art algorithms, with detection accuracy reaching approximately 85% across various scenarios.
Towards Blind and Low-Vision Accessibility of Lightweight VLMs and Custom LLM-Evals
Nov 13, 2025Abstract:Large Vision-Language Models (VLMs) excel at understanding and generating video descriptions but their high memory, computation, and deployment demands hinder practical use particularly for blind and low-vision (BLV) users who depend on detailed, context-aware descriptions. To study the effect of model size on accessibility-focused description quality, we evaluate SmolVLM2 variants with 500M and 2.2B parameters across two diverse datasets: AVCaps (outdoor), and Charades (indoor). In this work, we introduce two novel evaluation frameworks specifically designed for BLV accessibility assessment: the Multi-Context BLV Framework evaluating spatial orientation, social interaction, action events, and ambience contexts; and the Navigational Assistance Framework focusing on mobility-critical information. Additionally, we conduct a systematic evaluation of four different prompt design strategies and deploy both models on a smartphone, evaluating FP32 and INT8 precision variants to assess real-world performance constraints on resource-limited mobile devices.
EAD: An EEG Adapter for Automated Classification
May 29, 2025Abstract:While electroencephalography (EEG) has been a popular modality for neural decoding, it often involves task specific acquisition of the EEG data. This poses challenges for the development of a unified pipeline to learn embeddings for various EEG signal classification, which is often involved in various decoding tasks. Traditionally, EEG classification involves the step of signal preprocessing and the use of deep learning techniques, which are highly dependent on the number of EEG channels in each sample. However, the same pipeline cannot be applied even if the EEG data is collected for the same experiment but with different acquisition devices. This necessitates the development of a framework for learning EEG embeddings, which could be highly beneficial for tasks involving multiple EEG samples for the same task but with varying numbers of EEG channels. In this work, we propose EEG Adapter (EAD), a flexible framework compatible with any signal acquisition device. More specifically, we leverage a recent EEG foundational model with significant adaptations to learn robust representations from the EEG data for the classification task. We evaluate EAD on two publicly available datasets achieving state-of-the-art accuracies 99.33% and 92.31% on EEG-ImageNet and BrainLat respectively. This illustrates the effectiveness of the proposed framework across diverse EEG datasets containing two different perception tasks: stimulus and resting-state EEG signals. We also perform zero-shot EEG classification on EEG-ImageNet task to demonstrate the generalization capability of the proposed approach.
TractoGPT: A GPT architecture for White Matter Segmentation
Jan 26, 2025



Abstract:White matter bundle segmentation is crucial for studying brain structural connectivity, neurosurgical planning, and neurological disorders. White Matter Segmentation remains challenging due to structural similarity in streamlines, subject variability, symmetry in 2 hemispheres, etc. To address these challenges, we propose TractoGPT, a GPT-based architecture trained on streamline, cluster, and fusion data representations separately. TractoGPT is a fully-automatic method that generalizes across datasets and retains shape information of the white matter bundles. Experiments also show that TractoGPT outperforms state-of-the-art methods on average DICE, Overlap and Overreach scores. We use TractoInferno and 105HCP datasets and validate generalization across dataset.
MedSegDiffNCA: Diffusion Models With Neural Cellular Automata for Skin Lesion Segmentation
Jan 05, 2025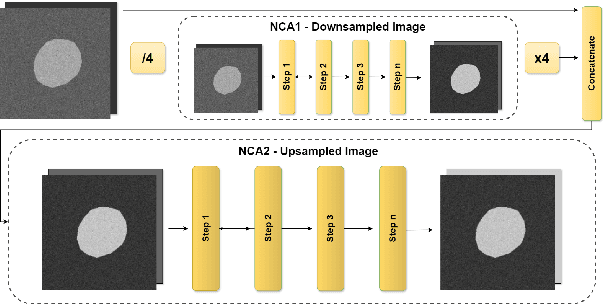
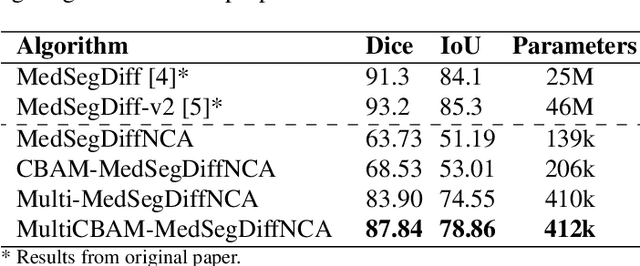
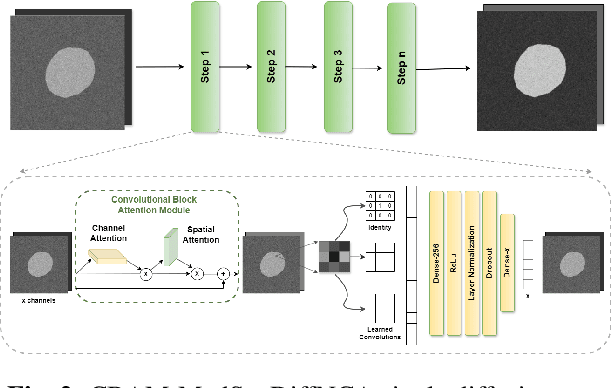

Abstract:Denoising Diffusion Models (DDMs) are widely used for high-quality image generation and medical image segmentation but often rely on Unet-based architectures, leading to high computational overhead, especially with high-resolution images. This work proposes three NCA-based improvements for diffusion-based medical image segmentation. First, Multi-MedSegDiffNCA uses a multilevel NCA framework to refine rough noise estimates generated by lower level NCA models. Second, CBAM-MedSegDiffNCA incorporates channel and spatial attention for improved segmentation. Third, MultiCBAM-MedSegDiffNCA combines these methods with a new RGB channel loss for semantic guidance. Evaluations on Lesion segmentation show that MultiCBAM-MedSegDiffNCA matches Unet-based model performance with dice score of 87.84% while using 60-110 times fewer parameters, offering a more efficient solution for low resource medical settings.
TractoEmbed: Modular Multi-level Embedding framework for white matter tract segmentation
Nov 12, 2024
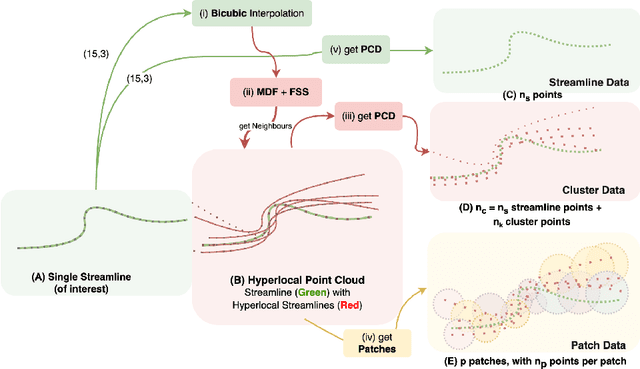
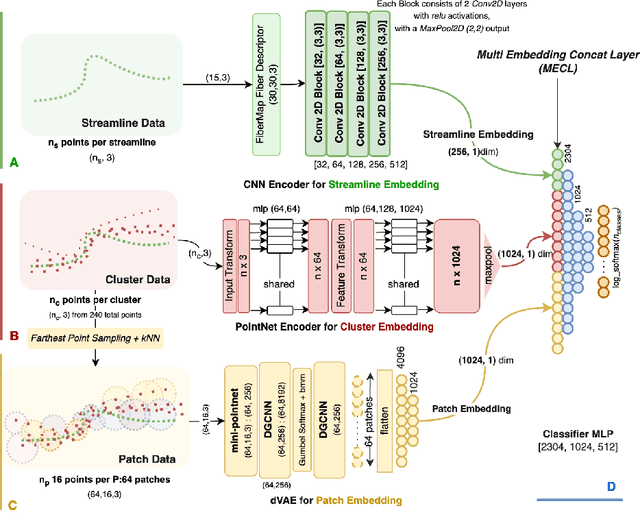
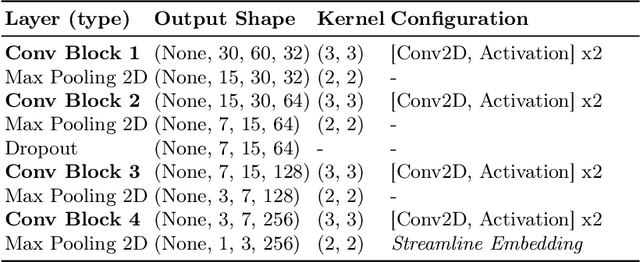
Abstract:White matter tract segmentation is crucial for studying brain structural connectivity and neurosurgical planning. However, segmentation remains challenging due to issues like class imbalance between major and minor tracts, structural similarity, subject variability, symmetric streamlines between hemispheres etc. To address these challenges, we propose TractoEmbed, a modular multi-level embedding framework, that encodes localized representations through learning tasks in respective encoders. In this paper, TractoEmbed introduces a novel hierarchical streamline data representation that captures maximum spatial information at each level i.e. individual streamlines, clusters, and patches. Experiments show that TractoEmbed outperforms state-of-the-art methods in white matter tract segmentation across different datasets, and spanning various age groups. The modular framework directly allows the integration of additional embeddings in future works.
Tract-RLFormer: A Tract-Specific RL policy based Decoder-only Transformer Network
Nov 08, 2024Abstract:Fiber tractography is a cornerstone of neuroimaging, enabling the detailed mapping of the brain's white matter pathways through diffusion MRI. This is crucial for understanding brain connectivity and function, making it a valuable tool in neurological applications. Despite its importance, tractography faces challenges due to its complexity and susceptibility to false positives, misrepresenting vital pathways. To address these issues, recent strategies have shifted towards deep learning, utilizing supervised learning, which depends on precise ground truth, or reinforcement learning, which operates without it. In this work, we propose Tract-RLFormer, a network utilizing both supervised and reinforcement learning, in a two-stage policy refinement process that markedly improves the accuracy and generalizability across various data-sets. By employing a tract-specific approach, our network directly delineates the tracts of interest, bypassing the traditional segmentation process. Through rigorous validation on datasets such as TractoInferno, HCP, and ISMRM-2015, our methodology demonstrates a leap forward in tractography, showcasing its ability to accurately map the brain's white matter tracts.
Unified Anomaly Detection methods on Edge Device using Knowledge Distillation and Quantization
Jul 03, 2024



Abstract:With the rapid advances in deep learning and smart manufacturing in Industry 4.0, there is an imperative for high-throughput, high-performance, and fully integrated visual inspection systems. Most anomaly detection approaches using defect detection datasets, such as MVTec AD, employ one-class models that require fitting separate models for each class. On the contrary, unified models eliminate the need for fitting separate models for each class and significantly reduce cost and memory requirements. Thus, in this work, we experiment with considering a unified multi-class setup. Our experimental study shows that multi-class models perform at par with one-class models for the standard MVTec AD dataset. Hence, this indicates that there may not be a need to learn separate object/class-wise models when the object classes are significantly different from each other, as is the case of the dataset considered. Furthermore, we have deployed three different unified lightweight architectures on the CPU and an edge device (NVIDIA Jetson Xavier NX). We analyze the quantized multi-class anomaly detection models in terms of latency and memory requirements for deployment on the edge device while comparing quantization-aware training (QAT) and post-training quantization (PTQ) for performance at different precision widths. In addition, we explored two different methods of calibration required in post-training scenarios and show that one of them performs notably better, highlighting its importance for unsupervised tasks. Due to quantization, the performance drop in PTQ is further compensated by QAT, which yields at par performance with the original 32-bit Floating point in two of the models considered.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge