Chirag Ahuja
TractRLFusion: A GPT-Based Multi-Critic Policy Fusion Framework for Fiber Tractography
Jan 20, 2026Abstract:Tractography plays a pivotal role in the non-invasive reconstruction of white matter fiber pathways, providing vital information on brain connectivity and supporting precise neurosurgical planning. Although traditional methods relied mainly on classical deterministic and probabilistic approaches, recent progress has benefited from supervised deep learning (DL) and deep reinforcement learning (DRL) to improve tract reconstruction. A persistent challenge in tractography is accurately reconstructing white matter tracts while minimizing spurious connections. To address this, we propose TractRLFusion, a novel GPT-based policy fusion framework that integrates multiple RL policies through a data-driven fusion strategy. Our method employs a two-stage training data selection process for effective policy fusion, followed by a multi-critic fine-tuning phase to enhance robustness and generalization. Experiments on HCP, ISMRM, and TractoInferno datasets demonstrate that TractRLFusion outperforms individual RL policies as well as state-of-the-art classical and DRL methods in accuracy and anatomical reliability.
TrackletGPT: A Language-like GPT Framework for White Matter Tract Segmentation
Jan 20, 2026Abstract:White Matter Tract Segmentation is imperative for studying brain structural connectivity, neurological disorders and neurosurgery. This task remains complex, as tracts differ among themselves, across subjects and conditions, yet have similar 3D structure across hemispheres and subjects. To address these challenges, we propose TrackletGPT, a language-like GPT framework which reintroduces sequential information in tokens using tracklets. TrackletGPT generalises seamlessly across datasets, is fully automatic, and encodes granular sub-streamline segments, Tracklets, scaling and refining GPT models in Tractography Segmentation. Based on our experiments, TrackletGPT outperforms state-of-the-art methods on average DICE, Overlap and Overreach scores on TractoInferno and HCP datasets, even on inter-dataset experiments.
TractoGPT: A GPT architecture for White Matter Segmentation
Jan 26, 2025



Abstract:White matter bundle segmentation is crucial for studying brain structural connectivity, neurosurgical planning, and neurological disorders. White Matter Segmentation remains challenging due to structural similarity in streamlines, subject variability, symmetry in 2 hemispheres, etc. To address these challenges, we propose TractoGPT, a GPT-based architecture trained on streamline, cluster, and fusion data representations separately. TractoGPT is a fully-automatic method that generalizes across datasets and retains shape information of the white matter bundles. Experiments also show that TractoGPT outperforms state-of-the-art methods on average DICE, Overlap and Overreach scores. We use TractoInferno and 105HCP datasets and validate generalization across dataset.
TractoEmbed: Modular Multi-level Embedding framework for white matter tract segmentation
Nov 12, 2024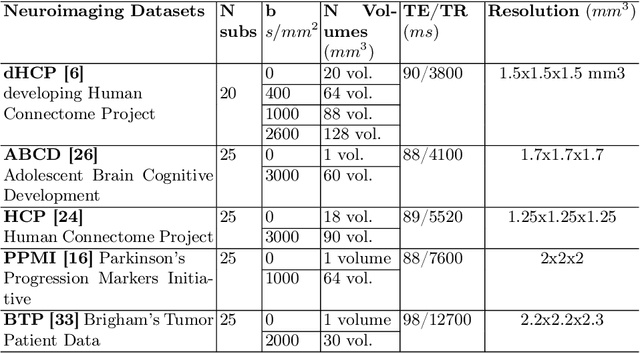
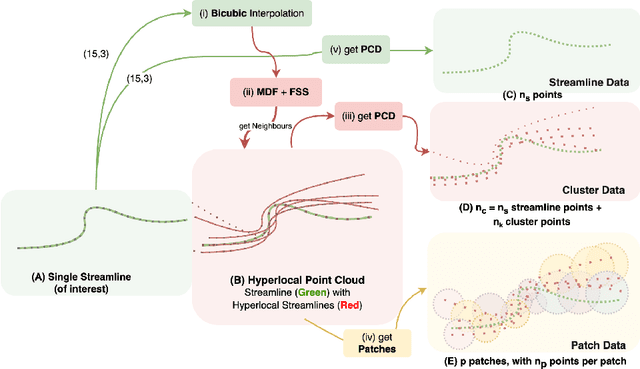
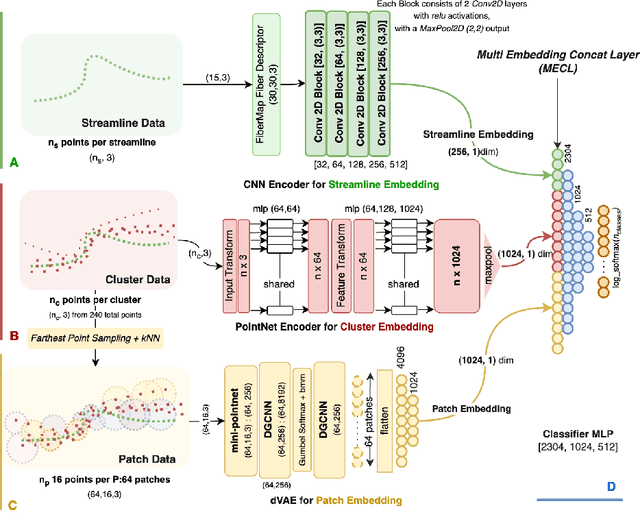
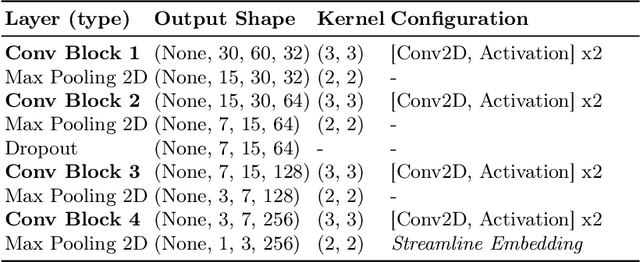
Abstract:White matter tract segmentation is crucial for studying brain structural connectivity and neurosurgical planning. However, segmentation remains challenging due to issues like class imbalance between major and minor tracts, structural similarity, subject variability, symmetric streamlines between hemispheres etc. To address these challenges, we propose TractoEmbed, a modular multi-level embedding framework, that encodes localized representations through learning tasks in respective encoders. In this paper, TractoEmbed introduces a novel hierarchical streamline data representation that captures maximum spatial information at each level i.e. individual streamlines, clusters, and patches. Experiments show that TractoEmbed outperforms state-of-the-art methods in white matter tract segmentation across different datasets, and spanning various age groups. The modular framework directly allows the integration of additional embeddings in future works.
Tract-RLFormer: A Tract-Specific RL policy based Decoder-only Transformer Network
Nov 08, 2024Abstract:Fiber tractography is a cornerstone of neuroimaging, enabling the detailed mapping of the brain's white matter pathways through diffusion MRI. This is crucial for understanding brain connectivity and function, making it a valuable tool in neurological applications. Despite its importance, tractography faces challenges due to its complexity and susceptibility to false positives, misrepresenting vital pathways. To address these issues, recent strategies have shifted towards deep learning, utilizing supervised learning, which depends on precise ground truth, or reinforcement learning, which operates without it. In this work, we propose Tract-RLFormer, a network utilizing both supervised and reinforcement learning, in a two-stage policy refinement process that markedly improves the accuracy and generalizability across various data-sets. By employing a tract-specific approach, our network directly delineates the tracts of interest, bypassing the traditional segmentation process. Through rigorous validation on datasets such as TractoInferno, HCP, and ISMRM-2015, our methodology demonstrates a leap forward in tractography, showcasing its ability to accurately map the brain's white matter tracts.
An Automatic Method for Complete Brain Matter Segmentation from Multislice CT scan
Oct 22, 2018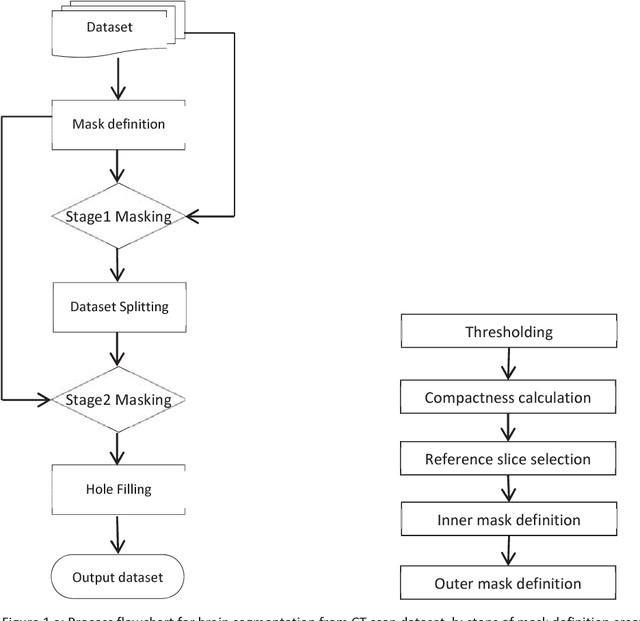
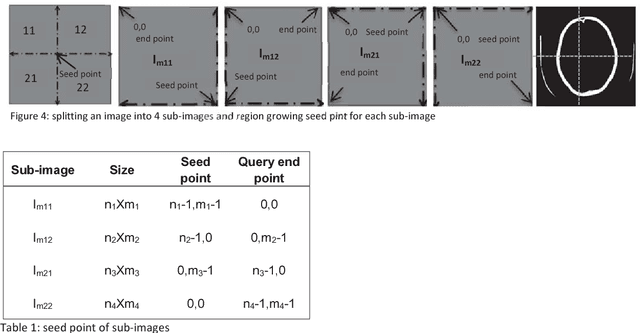
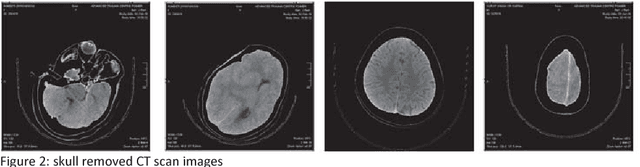
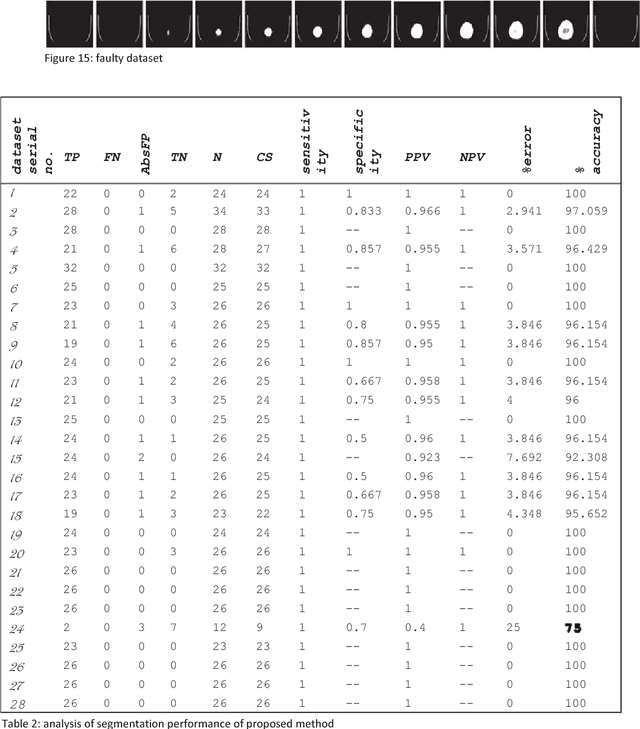
Abstract:Computed tomography imaging is well accepted for its imaging speed, image contrast & resolution and cost. Thus it has wide use in detection and diagnosis of brain diseases. But unfortunately reported works on CT segmentation is not very significant. In this paper, a robust automatic segmentation system is presented which is capable of segment complete brain matter from CT slices, without any lose in information. The proposed method is simple, fast, accurate and completely automatic. It can handle multislice CT scan in single run. From a given multislice CT dataset, one slice is selected automatically to form masks for segmentation. Two types of masks are created to handle nasal slices in a better way. Masks are created from selected reference slice using automatic seed point selection and region growing technique. One mask is designed for brain matter and another includes the skull of the reference slice. This second mask is used as global reference mask for all slices whereas the brain matter mask is implemented on only adjacent slices and continuously modified for better segmentation. Slices in given dataset are divided into two batches, before reference slice and after reference slice. Each batch segmented separately. Successive propagation of brain matter mask has demonstrated very high potential in reported segmentation. Presented result shows highest sensitivity and more than 96% accuracy in all cases. Resulted segmented images can be used for any brain disease diagnosis or further image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge