Ilkay Oksuz
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
HyperCMR: Enhanced Multi-Contrast CMR Reconstruction with Eagle Loss
Oct 04, 2024Abstract:Accelerating image acquisition for cardiac magnetic resonance imaging (CMRI) is a critical task. CMRxRecon2024 challenge aims to set the state of the art for multi-contrast CMR reconstruction. This paper presents HyperCMR, a novel framework designed to accelerate the reconstruction of multi-contrast cardiac magnetic resonance (CMR) images. HyperCMR enhances the existing PromptMR model by incorporating advanced loss functions, notably the innovative Eagle Loss, which is specifically designed to recover missing high-frequency information in undersampled k-space. Extensive experiments conducted on the CMRxRecon2024 challenge dataset demonstrate that HyperCMR consistently outperforms the baseline across multiple evaluation metrics, achieving superior SSIM and PSNR scores.
Mammographic Breast Positioning Assessment via Deep Learning
Jul 15, 2024



Abstract:Breast cancer remains a leading cause of cancer-related deaths among women worldwide, with mammography screening as the most effective method for the early detection. Ensuring proper positioning in mammography is critical, as poor positioning can lead to diagnostic errors, increased patient stress, and higher costs due to recalls. Despite advancements in deep learning (DL) for breast cancer diagnostics, limited focus has been given to evaluating mammography positioning. This paper introduces a novel DL methodology to quantitatively assess mammogram positioning quality, specifically in mediolateral oblique (MLO) views using attention and coordinate convolution modules. Our method identifies key anatomical landmarks, such as the nipple and pectoralis muscle, and automatically draws a posterior nipple line (PNL), offering robust and inherently explainable alternative to well-known classification and regression-based approaches. We compare the performance of proposed methodology with various regression and classification-based models. The CoordAtt UNet model achieved the highest accuracy of 88.63% $\pm$ 2.84 and specificity of 90.25% $\pm$ 4.04, along with a noteworthy sensitivity of 86.04% $\pm$ 3.41. In landmark detection, the same model also recorded the lowest mean errors in key anatomical points and the smallest angular error of 2.42 degrees. Our results indicate that models incorporating attention mechanisms and CoordConv module increase the accuracy in classifying breast positioning quality and detecting anatomical landmarks. Furthermore, we make the labels and source codes available to the community to initiate an open research area for mammography, accessible at https://github.com/tanyelai/deep-breast-positioning.
Explainable Image Quality Assessment for Medical Imaging
Mar 25, 2023



Abstract:Medical image quality assessment is an important aspect of image acquisition, as poor-quality images may lead to misdiagnosis. Manual labelling of image quality is a tedious task for population studies and can lead to misleading results. While much research has been done on automated analysis of image quality to address this issue, relatively little work has been done to explain the methodologies. In this work, we propose an explainable image quality assessment system and validate our idea on two different objectives which are foreign object detection on Chest X-Rays (Object-CXR) and Left Ventricular Outflow Tract (LVOT) detection on Cardiac Magnetic Resonance (CMR) volumes. We apply a variety of techniques to measure the faithfulness of the saliency detectors, and our explainable pipeline relies on NormGrad, an algorithm which can efficiently localise image quality issues with saliency maps of the classifier. We compare NormGrad with a range of saliency detection methods and illustrate its superior performance as a result of applying these methodologies for measuring the faithfulness of the saliency detectors. We see that NormGrad has significant gains over other saliency detectors by reaching a repeated Pointing Game score of 0.853 for Object-CXR and 0.611 for LVOT datasets.
Semi-Supervised Segmentation of Multi-vendor and Multi-center Cardiac MRI using Histogram Matching
Feb 22, 2023Abstract:Automatic segmentation of the heart cavity is an essential task for the diagnosis of cardiac diseases. In this paper, we propose a semi-supervised segmentation setup for leveraging unlabeled data to segment Left-ventricle, Right-ventricle, and Myocardium. We utilize an enhanced version of residual U-Net architecture on a large-scale cardiac MRI dataset. Handling the class imbalanced data issue using dice loss, the enhanced supervised model is able to achieve better dice scores in comparison with a vanilla U-Net model. We applied several augmentation techniques including histogram matching to increase the performance of our model in other domains. Also, we introduce a simple but efficient semi-supervised segmentation method to improve segmentation results without the need for large labeled data. Finally, we applied our method on two benchmark datasets, STACOM2018, and M\&Ms 2020 challenges, to show the potency of the proposed model. The effectiveness of our proposed model is demonstrated by the quantitative results. The model achieves average dice scores of 0.921, 0.926, and 0.891 for Left-ventricle, Right-ventricle, and Myocardium respectively.
* 5 pages, 8 figures, IEEE conference published paper
Prostate Lesion Estimation using Prostate Masks from Biparametric MRI
Jan 11, 2023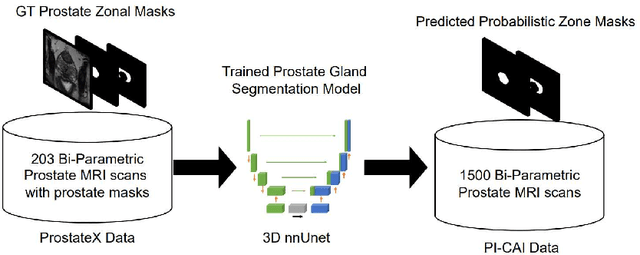
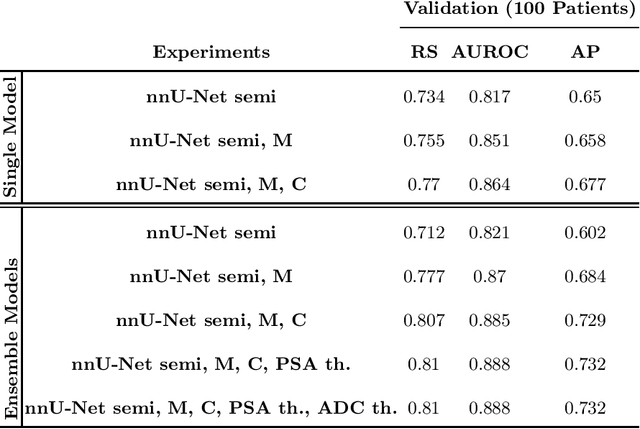
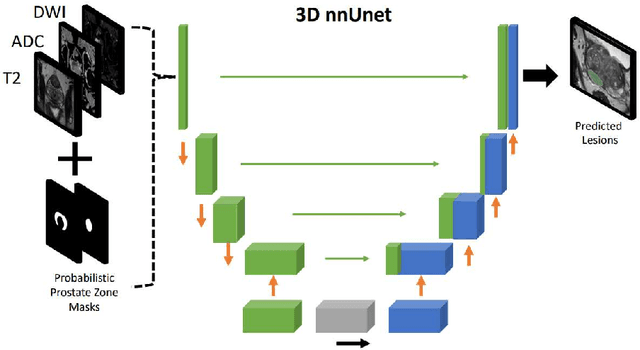
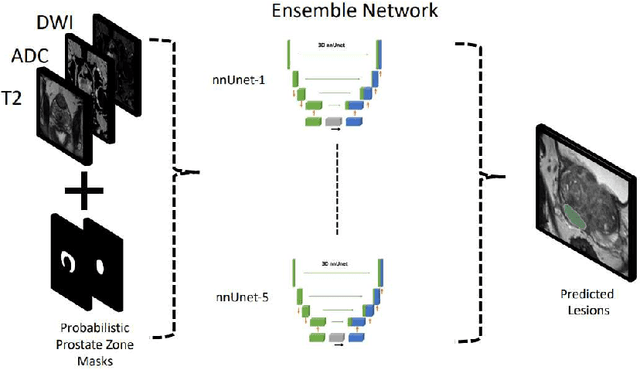
Abstract:Biparametric MRI has emerged as an alternative to multiparametric prostate MRI, which eliminates the need for the potential harms to the patient due to the contrast medium. One major issue with biparametric MRI is difficulty to detect clinically significant prostate cancer (csPCA). Deep learning algorithms have emerged as an alternative solution to detect csPCA in cohort studies. We present a workflow which predicts csPCA on biparametric prostate MRI PI-CAI 2022 Challenge with over 10,000 carefully-curated prostate MRI exams. We propose to to segment the prostate gland first to the central gland (transition + central zone) and the peripheral gland. Then we utilize these predcitions in combination with T2, ADC and DWI images to train an ensemble nnU-Net model. Finally, we utilize clinical indices PSA and ADC intensity distributions of lesion regions to reduce the false positives. Our method achieves top results on open-validation stage with a AUROC of 0.888 and AP of 0.732.
Detecting respiratory motion artefacts for cardiovascular MRIs to ensure high-quality segmentation
Sep 20, 2022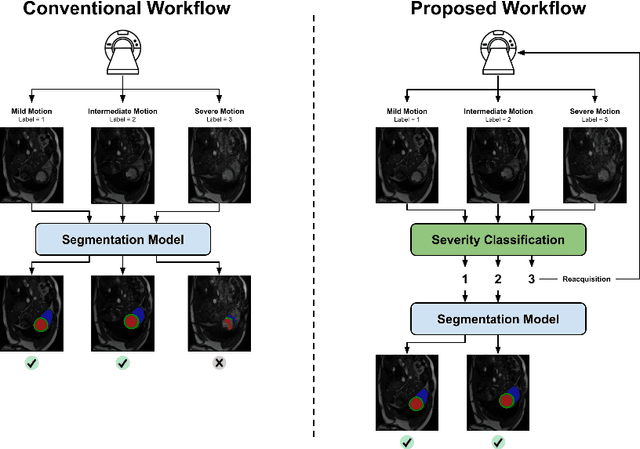
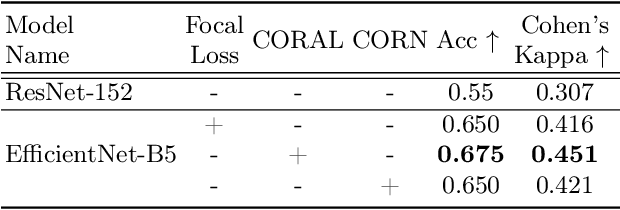
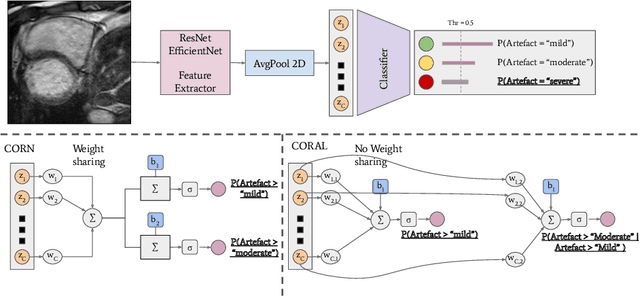
Abstract:While machine learning approaches perform well on their training domain, they generally tend to fail in a real-world application. In cardiovascular magnetic resonance imaging (CMR), respiratory motion represents a major challenge in terms of acquisition quality and therefore subsequent analysis and final diagnosis. We present a workflow which predicts a severity score for respiratory motion in CMR for the CMRxMotion challenge 2022. This is an important tool for technicians to immediately provide feedback on the CMR quality during acquisition, as poor-quality images can directly be re-acquired while the patient is still available in the vicinity. Thus, our method ensures that the acquired CMR holds up to a specific quality standard before it is used for further diagnosis. Therefore, it enables an efficient base for proper diagnosis without having time and cost-intensive re-acquisitions in cases of severe motion artefacts. Combined with our segmentation model, this can help cardiologists and technicians in their daily routine by providing a complete pipeline to guarantee proper quality assessment and genuine segmentations for cardiovascular scans. The code base is available at https://github.com/MECLabTUDA/QA_med_data/tree/dev_QA_CMRxMotion.
Shifted Windows Transformers for Medical Image Quality Assessment
Aug 11, 2022



Abstract:To maintain a standard in a medical imaging study, images should have necessary image quality for potential diagnostic use. Although CNN-based approaches are used to assess the image quality, their performance can still be improved in terms of accuracy. In this work, we approach this problem by using Swin Transformer, which improves the poor-quality image classification performance that causes the degradation in medical image quality. We test our approach on Foreign Object Classification problem on Chest X-Rays (Object-CXR) and Left Ventricular Outflow Tract Classification problem on Cardiac MRI with a four-chamber view (LVOT). While we obtain a classification accuracy of 87.1% and 95.48% on the Object-CXR and LVOT datasets, our experimental results suggest that the use of Swin Transformer improves the Object-CXR classification performance while obtaining a comparable performance for the LVOT dataset. To the best of our knowledge, our study is the first vision transformer application for medical image quality assessment.
A Deep Learning-based Integrated Framework for Quality-aware Undersampled Cine Cardiac MRI Reconstruction and Analysis
May 02, 2022



Abstract:Cine cardiac magnetic resonance (CMR) imaging is considered the gold standard for cardiac function evaluation. However, cine CMR acquisition is inherently slow and in recent decades considerable effort has been put into accelerating scan times without compromising image quality or the accuracy of derived results. In this paper, we present a fully-automated, quality-controlled integrated framework for reconstruction, segmentation and downstream analysis of undersampled cine CMR data. The framework enables active acquisition of radial k-space data, in which acquisition can be stopped as soon as acquired data are sufficient to produce high quality reconstructions and segmentations. This results in reduced scan times and automated analysis, enabling robust and accurate estimation of functional biomarkers. To demonstrate the feasibility of the proposed approach, we perform realistic simulations of radial k-space acquisitions on a dataset of subjects from the UK Biobank and present results on in-vivo cine CMR k-space data collected from healthy subjects. The results demonstrate that our method can produce quality-controlled images in a mean scan time reduced from 12 to 4 seconds per slice, and that image quality is sufficient to allow clinically relevant parameters to be automatically estimated to within 5% mean absolute difference.
MyoPS: A Benchmark of Myocardial Pathology Segmentation Combining Three-Sequence Cardiac Magnetic Resonance Images
Jan 10, 2022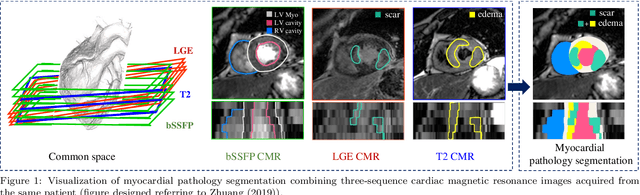
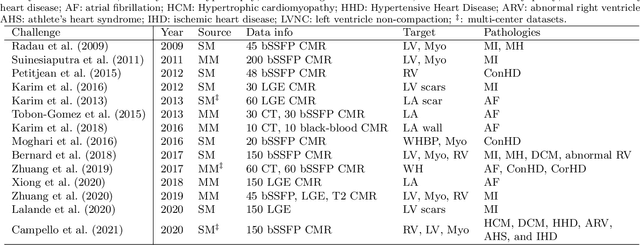
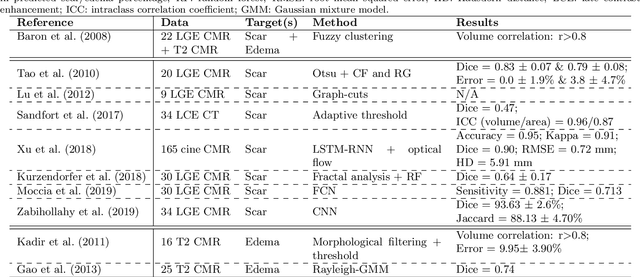
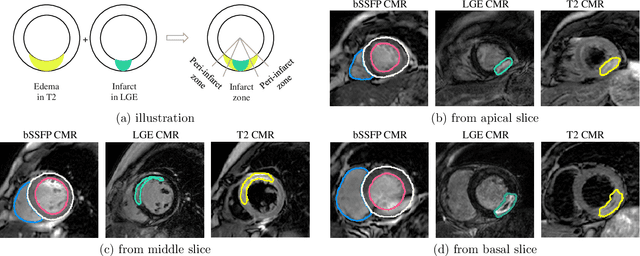
Abstract:Assessment of myocardial viability is essential in diagnosis and treatment management of patients suffering from myocardial infarction, and classification of pathology on myocardium is the key to this assessment. This work defines a new task of medical image analysis, i.e., to perform myocardial pathology segmentation (MyoPS) combining three-sequence cardiac magnetic resonance (CMR) images, which was first proposed in the MyoPS challenge, in conjunction with MICCAI 2020. The challenge provided 45 paired and pre-aligned CMR images, allowing algorithms to combine the complementary information from the three CMR sequences for pathology segmentation. In this article, we provide details of the challenge, survey the works from fifteen participants and interpret their methods according to five aspects, i.e., preprocessing, data augmentation, learning strategy, model architecture and post-processing. In addition, we analyze the results with respect to different factors, in order to examine the key obstacles and explore potential of solutions, as well as to provide a benchmark for future research. We conclude that while promising results have been reported, the research is still in the early stage, and more in-depth exploration is needed before a successful application to the clinics. Note that MyoPS data and evaluation tool continue to be publicly available upon registration via its homepage (www.sdspeople.fudan.edu.cn/zhuangxiahai/0/myops20/).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge