Esther Puyol-Antón
King's College London
Localising Shortcut Learning in Pixel Space via Ordinal Scoring Correlations for Attribution Representations (OSCAR)
Dec 21, 2025



Abstract:Deep neural networks often exploit shortcuts. These are spurious cues which are associated with output labels in the training data but are unrelated to task semantics. When the shortcut features are associated with sensitive attributes, shortcut learning can lead to biased model performance. Existing methods for localising and understanding shortcut learning are mostly based upon qualitative, image-level inspection and assume cues are human-visible, limiting their use in domains such as medical imaging. We introduce OSCAR (Ordinal Scoring Correlations for Attribution Representations), a model-agnostic framework for quantifying shortcut learning and localising shortcut features. OSCAR converts image-level task attribution maps into dataset-level rank profiles of image regions and compares them across three models: a balanced baseline model (BA), a test model (TS), and a sensitive attribute predictor (SA). By computing pairwise, partial, and deviation-based correlations on these rank profiles, we produce a set of quantitative metrics that characterise the degree of shortcut reliance for TS, together with a ranking of image-level regions that contribute most to it. Experiments on CelebA, CheXpert, and ADNI show that our correlations are (i) stable across seeds and partitions, (ii) sensitive to the level of association between shortcut features and output labels in the training data, and (iii) able to distinguish localised from diffuse shortcut features. As an illustration of the utility of our method, we show how worst-group performance disparities can be reduced using a simple test-time attenuation approach based on the identified shortcut regions. OSCAR provides a lightweight, pixel-space audit that yields statistical decision rules and spatial maps, enabling users to test, localise, and mitigate shortcut reliance. The code is available at https://github.com/acharaakshit/oscar
DeepSPV: An Interpretable Deep Learning Pipeline for 3D Spleen Volume Estimation from 2D Ultrasound Images
Nov 17, 2024



Abstract:Splenomegaly, the enlargement of the spleen, is an important clinical indicator for various associated medical conditions, such as sickle cell disease (SCD). Spleen length measured from 2D ultrasound is the most widely used metric for characterising spleen size. However, it is still considered a surrogate measure, and spleen volume remains the gold standard for assessing spleen size. Accurate spleen volume measurement typically requires 3D imaging modalities, such as computed tomography or magnetic resonance imaging, but these are not widely available, especially in the Global South which has a high prevalence of SCD. In this work, we introduce a deep learning pipeline, DeepSPV, for precise spleen volume estimation from single or dual 2D ultrasound images. The pipeline involves a segmentation network and a variational autoencoder for learning low-dimensional representations from the estimated segmentations. We investigate three approaches for spleen volume estimation and our best model achieves 86.62%/92.5% mean relative volume accuracy (MRVA) under single-view/dual-view settings, surpassing the performance of human experts. In addition, the pipeline can provide confidence intervals for the volume estimates as well as offering benefits in terms of interpretability, which further support clinicians in decision-making when identifying splenomegaly. We evaluate the full pipeline using a highly realistic synthetic dataset generated by a diffusion model, achieving an overall MRVA of 83.0% from a single 2D ultrasound image. Our proposed DeepSPV is the first work to use deep learning to estimate 3D spleen volume from 2D ultrasound images and can be seamlessly integrated into the current clinical workflow for spleen assessment.
Improving the Scan-rescan Precision of AI-based CMR Biomarker Estimation
Aug 21, 2024Abstract:Quantification of cardiac biomarkers from cine cardiovascular magnetic resonance (CMR) data using deep learning (DL) methods offers many advantages, such as increased accuracy and faster analysis. However, only a few studies have focused on the scan-rescan precision of the biomarker estimates, which is important for reproducibility and longitudinal analysis. Here, we propose a cardiac biomarker estimation pipeline that not only focuses on achieving high segmentation accuracy but also on improving the scan-rescan precision of the computed biomarkers, namely left and right ventricular ejection fraction, and left ventricular myocardial mass. We evaluate two approaches to improve the apical-basal resolution of the segmentations used for estimating the biomarkers: one based on image interpolation and one based on segmentation interpolation. Using a database comprising scan-rescan cine CMR data acquired from 92 subjects, we compare the performance of these two methods against ground truth (GT) segmentations and DL segmentations obtained before interpolation (baseline). The results demonstrate that both the image-based and segmentation-based interpolation methods were able to narrow Bland-Altman scan-rescan confidence intervals for all biomarkers compared to the GT and baseline performances. Our findings highlight the importance of focusing not only on segmentation accuracy but also on the consistency of biomarkers across repeated scans, which is crucial for longitudinal analysis of cardiac function.
Improved 3D Whole Heart Geometry from Sparse CMR Slices
Aug 14, 2024Abstract:Cardiac magnetic resonance (CMR) imaging and computed tomography (CT) are two common non-invasive imaging methods for assessing patients with cardiovascular disease. CMR typically acquires multiple sparse 2D slices, with unavoidable respiratory motion artefacts between slices, whereas CT acquires isotropic dense data but uses ionising radiation. In this study, we explore the combination of Slice Shifting Algorithm (SSA), Spatial Transformer Network (STN), and Label Transformer Network (LTN) to: 1) correct respiratory motion between segmented slices, and 2) transform sparse segmentation data into dense segmentation. All combinations were validated using synthetic motion-corrupted CMR slice segmentation generated from CT in 1699 cases, where the dense CT serves as the ground truth. In 199 testing cases, SSA-LTN achieved the best results for Dice score and Huasdorff distance (94.0% and 4.7 mm respectively, average over 5 labels) but gave topological errors in 8 cases. STN was effective as a plug-in tool for correcting all topological errors with minimal impact on overall performance (93.5% and 5.0 mm respectively). SSA also proves to be a valuable plug-in tool, enhancing performance over both STN-based and LTN-based models. The code for these different combinations is available at https://github.com/XESchong/STACOM2024.
Improving Deep Learning Model Calibration for Cardiac Applications using Deterministic Uncertainty Networks and Uncertainty-aware Training
May 10, 2024



Abstract:Improving calibration performance in deep learning (DL) classification models is important when planning the use of DL in a decision-support setting. In such a scenario, a confident wrong prediction could lead to a lack of trust and/or harm in a high-risk application. We evaluate the impact on accuracy and calibration of two types of approach that aim to improve DL classification model calibration: deterministic uncertainty methods (DUM) and uncertainty-aware training. Specifically, we test the performance of three DUMs and two uncertainty-aware training approaches as well as their combinations. To evaluate their utility, we use two realistic clinical applications from the field of cardiac imaging: artefact detection from phase contrast cardiac magnetic resonance (CMR) and disease diagnosis from the public ACDC CMR dataset. Our results indicate that both DUMs and uncertainty-aware training can improve both accuracy and calibration in both of our applications, with DUMs generally offering the best improvements. We also investigate the combination of the two approaches, resulting in a novel deterministic uncertainty-aware training approach. This provides further improvements for some combinations of DUMs and uncertainty-aware training approaches.
Label Dropout: Improved Deep Learning Echocardiography Segmentation Using Multiple Datasets With Domain Shift and Partial Labelling
Mar 12, 2024Abstract:Echocardiography (echo) is the first imaging modality used when assessing cardiac function. The measurement of functional biomarkers from echo relies upon the segmentation of cardiac structures and deep learning models have been proposed to automate the segmentation process. However, in order to translate these tools to widespread clinical use it is important that the segmentation models are robust to a wide variety of images (e.g. acquired from different scanners, by operators with different levels of expertise etc.). To achieve this level of robustness it is necessary that the models are trained with multiple diverse datasets. A significant challenge faced when training with multiple diverse datasets is the variation in label presence, i.e. the combined data are often partially-labelled. Adaptations of the cross entropy loss function have been proposed to deal with partially labelled data. In this paper we show that training naively with such a loss function and multiple diverse datasets can lead to a form of shortcut learning, where the model associates label presence with domain characteristics, leading to a drop in performance. To address this problem, we propose a novel label dropout scheme to break the link between domain characteristics and the presence or absence of labels. We demonstrate that label dropout improves echo segmentation Dice score by 62% and 25% on two cardiac structures when training using multiple diverse partially labelled datasets.
Bias in Unsupervised Anomaly Detection in Brain MRI
Aug 26, 2023



Abstract:Unsupervised anomaly detection methods offer a promising and flexible alternative to supervised approaches, holding the potential to revolutionize medical scan analysis and enhance diagnostic performance. In the current landscape, it is commonly assumed that differences between a test case and the training distribution are attributed solely to pathological conditions, implying that any disparity indicates an anomaly. However, the presence of other potential sources of distributional shift, including scanner, age, sex, or race, is frequently overlooked. These shifts can significantly impact the accuracy of the anomaly detection task. Prominent instances of such failures have sparked concerns regarding the bias, credibility, and fairness of anomaly detection. This work presents a novel analysis of biases in unsupervised anomaly detection. By examining potential non-pathological distributional shifts between the training and testing distributions, we shed light on the extent of these biases and their influence on anomaly detection results. Moreover, this study examines the algorithmic limitations that arise due to biases, providing valuable insights into the challenges encountered by anomaly detection algorithms in accurately learning and capturing the entire range of variability present in the normative distribution. Through this analysis, we aim to enhance the understanding of these biases and pave the way for future improvements in the field. Here, we specifically investigate Alzheimer's disease detection from brain MR imaging as a case study, revealing significant biases related to sex, race, and scanner variations that substantially impact the results. These findings align with the broader goal of improving the reliability, fairness, and effectiveness of anomaly detection in medical imaging.
An investigation into the impact of deep learning model choice on sex and race bias in cardiac MR segmentation
Aug 25, 2023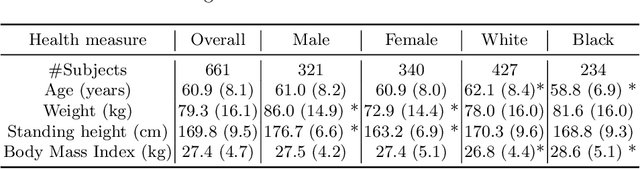



Abstract:In medical imaging, artificial intelligence (AI) is increasingly being used to automate routine tasks. However, these algorithms can exhibit and exacerbate biases which lead to disparate performances between protected groups. We investigate the impact of model choice on how imbalances in subject sex and race in training datasets affect AI-based cine cardiac magnetic resonance image segmentation. We evaluate three convolutional neural network-based models and one vision transformer model. We find significant sex bias in three of the four models and racial bias in all of the models. However, the severity and nature of the bias varies between the models, highlighting the importance of model choice when attempting to train fair AI-based segmentation models for medical imaging tasks.
Automated Quality Controlled Analysis of 2D Phase Contrast Cardiovascular Magnetic Resonance Imaging
Sep 28, 2022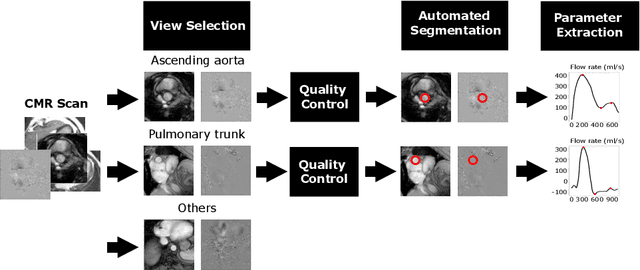
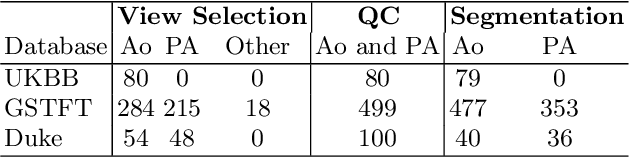
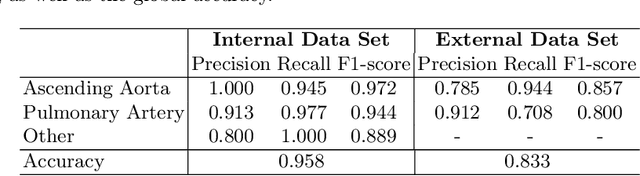
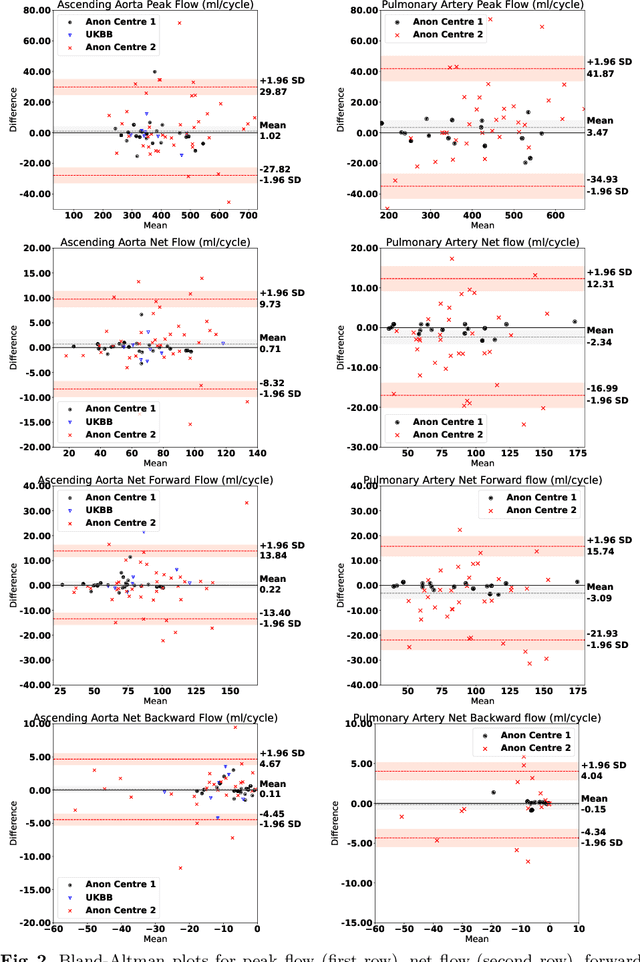
Abstract:Flow analysis carried out using phase contrast cardiac magnetic resonance imaging (PC-CMR) enables the quantification of important parameters that are used in the assessment of cardiovascular function. An essential part of this analysis is the identification of the correct CMR views and quality control (QC) to detect artefacts that could affect the flow quantification. We propose a novel deep learning based framework for the fully-automated analysis of flow from full CMR scans that first carries out these view selection and QC steps using two sequential convolutional neural networks, followed by automatic aorta and pulmonary artery segmentation to enable the quantification of key flow parameters. Accuracy values of 0.958 and 0.914 were obtained for view classification and QC, respectively. For segmentation, Dice scores were $>$0.969 and the Bland-Altman plots indicated excellent agreement between manual and automatic peak flow values. In addition, we tested our pipeline on an external validation data set, with results indicating good robustness of the pipeline. This work was carried out using multivendor clinical data consisting of 986 cases, indicating the potential for the use of this pipeline in a clinical setting.
Large-scale, multi-centre, multi-disease validation of an AI clinical tool for cine CMR analysis
Jun 15, 2022



Abstract:INTRODUCTION: Artificial intelligence (AI) has the potential to facilitate the automation of CMR analysis for biomarker extraction. However, most AI algorithms are trained on a specific input domain (e.g., single scanner vendor or hospital-tailored imaging protocol) and lack the robustness to perform optimally when applied to CMR data from other input domains. METHODS: Our proposed framework consists of an AI-based algorithm for biventricular segmentation of short-axis images, followed by a post-analysis quality control to detect erroneous results. The segmentation algorithm was trained on a large dataset of clinical CMR scans from two NHS hospitals (n=2793) and validated on additional cases from this dataset (n=441) and on five external datasets (n=6808). The validation data included CMR scans of patients with a range of diseases acquired at 12 different centres using CMR scanners from all major vendors. RESULTS: Our method yielded median Dice scores over 87%, translating into median absolute errors in cardiac biomarkers within the range of inter-observer variability: <8.4mL (left ventricle), <9.2mL (right ventricle), <13.3g (left ventricular mass), and <5.9% (ejection fraction) across all datasets. Stratification of cases according to phenotypes of cardiac disease and scanner vendors showed good agreement. CONCLUSIONS: We show that our proposed tool, which combines a state-of-the-art AI algorithm trained on a large-scale multi-domain CMR dataset with a post-analysis quality control, allows us to robustly deal with routine clinical data from multiple centres, vendors, and cardiac diseases. This is a fundamental step for the clinical translation of AI algorithms. Moreover, our method yields a range of additional biomarkers of cardiac function (filling and ejection rates, regional wall motion, and strain) at no extra computational cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge