Emily Chan
Semantic Alignment of Unimodal Medical Text and Vision Representations
Mar 06, 2025Abstract:General-purpose AI models, particularly those designed for text and vision, demonstrate impressive versatility across a wide range of deep-learning tasks. However, they often underperform in specialised domains like medical imaging, where domain-specific solutions or alternative knowledge transfer approaches are typically required. Recent studies have noted that general-purpose models can exhibit similar latent spaces when processing semantically related data, although this alignment does not occur naturally. Building on this insight, it has been shown that applying a simple transformation - at most affine - estimated from a subset of semantically corresponding samples, known as anchors, enables model stitching across diverse training paradigms, architectures, and modalities. In this paper, we explore how semantic alignment - estimating transformations between anchors - can bridge general-purpose AI with specialised medical knowledge. Using multiple public chest X-ray datasets, we demonstrate that model stitching across model architectures allows general models to integrate domain-specific knowledge without additional training, leading to improved performance on medical tasks. Furthermore, we introduce a novel zero-shot classification approach for unimodal vision encoders that leverages semantic alignment across modalities. Our results show that our method not only outperforms general multimodal models but also approaches the performance levels of fully trained, medical-specific multimodal solutions
Unsupervised Analysis of Alzheimer's Disease Signatures using 3D Deformable Autoencoders
Jul 04, 2024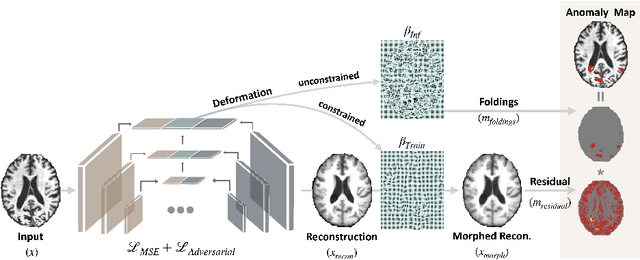
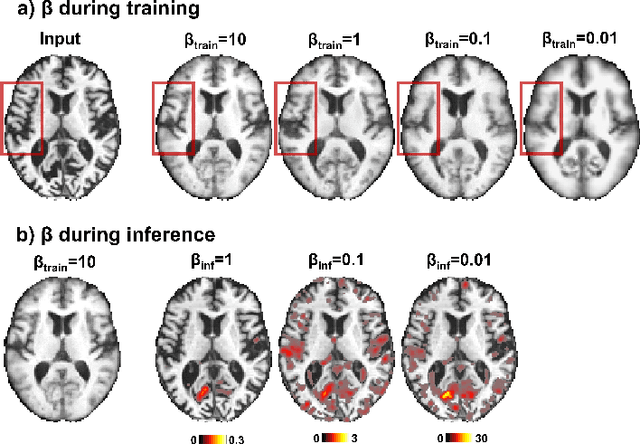
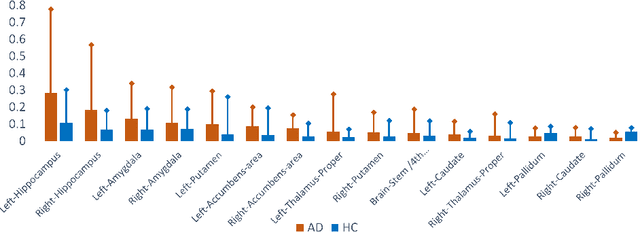
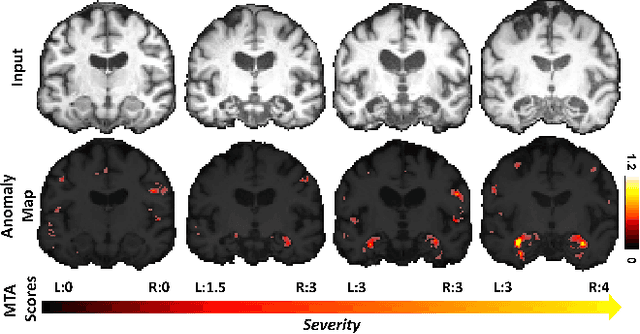
Abstract:With the increasing incidence of neurodegenerative diseases such as Alzheimer's Disease (AD), there is a need for further research that enhances detection and monitoring of the diseases. We present MORPHADE (Morphological Autoencoders for Alzheimer's Disease Detection), a novel unsupervised learning approach which uses deformations to allow the analysis of 3D T1-weighted brain images. To the best of our knowledge, this is the first use of deformations with deep unsupervised learning to not only detect, but also localize and assess the severity of structural changes in the brain due to AD. We obtain markedly higher anomaly scores in clinically important areas of the brain in subjects with AD compared to healthy controls, showcasing that our method is able to effectively locate AD-related atrophy. We additionally observe a visual correlation between the severity of atrophy highlighted in our anomaly maps and medial temporal lobe atrophy scores evaluated by a clinical expert. Finally, our method achieves an AUROC of 0.80 in detecting AD, out-performing several supervised and unsupervised baselines. We believe our framework shows promise as a tool towards improved understanding, monitoring and detection of AD. To support further research and application, we have made our code publicly available at github.com/ci-ber/MORPHADE.
Addressing Deep Learning Model Calibration Using Evidential Neural Networks and Uncertainty-Aware Training
Jan 30, 2023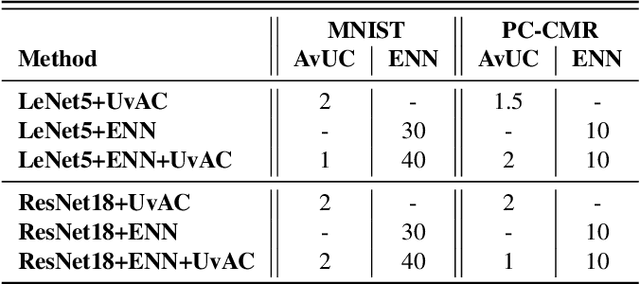

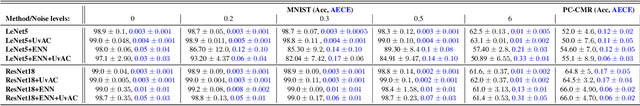
Abstract:In terms of accuracy, deep learning (DL) models have had considerable success in classification problems for medical imaging applications. However, it is well-known that the outputs of such models, which typically utilise the SoftMax function in the final classification layer can be over-confident, i.e. they are poorly calibrated. Two competing solutions to this problem have been proposed: uncertainty-aware training and evidential neural networks (ENNs). In this paper, we perform an investigation into the improvements to model calibration that can be achieved by each of these approaches individually, and their combination. We perform experiments on two classification tasks: a simpler MNIST digit classification task and a more complex and realistic medical imaging artefact detection task using Phase Contrast Cardiac Magnetic Resonance images. The experimental results demonstrate that model calibration can suffer when the task becomes challenging enough to require a higher-capacity model. However, in our complex artefact detection task, we saw an improvement in calibration for both a low and higher-capacity model when implementing both the ENN and uncertainty-aware training together, indicating that this approach can offer a promising way to improve calibration in such settings. The findings highlight the potential use of these approaches to improve model calibration in a complex application, which would in turn improve clinician trust in DL models.
Automated Quality Controlled Analysis of 2D Phase Contrast Cardiovascular Magnetic Resonance Imaging
Sep 28, 2022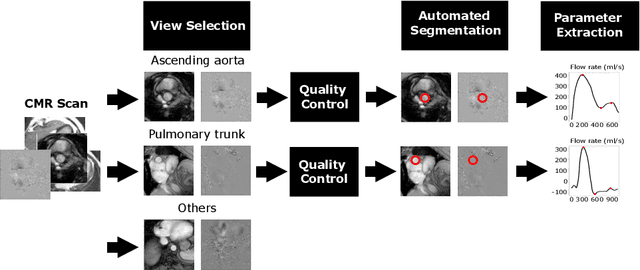
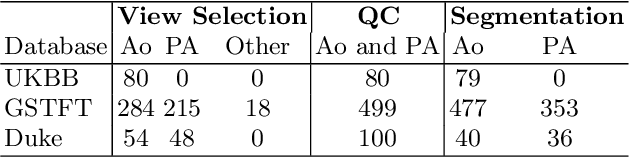
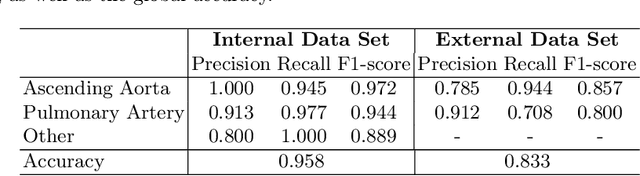
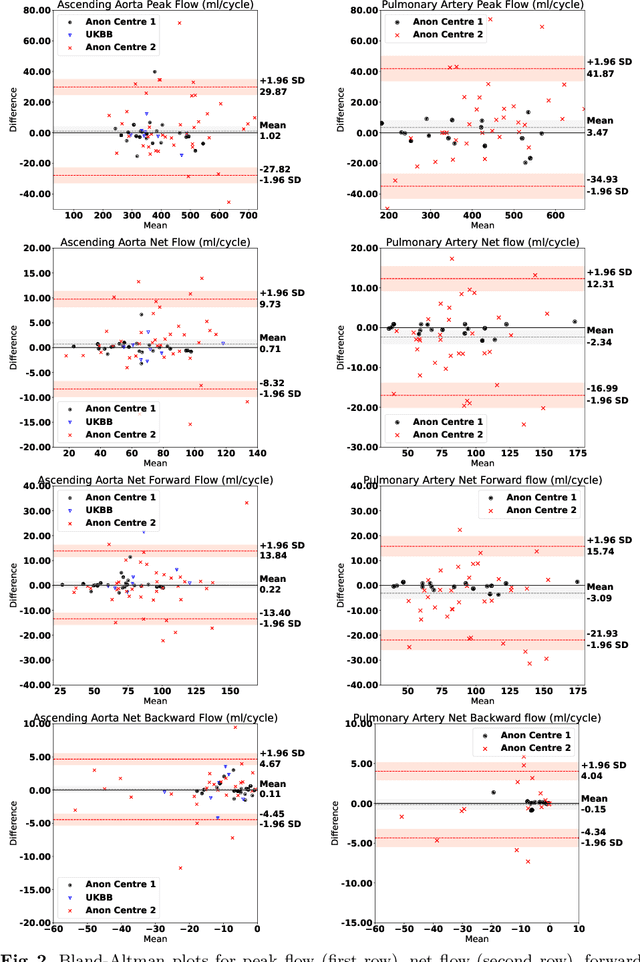
Abstract:Flow analysis carried out using phase contrast cardiac magnetic resonance imaging (PC-CMR) enables the quantification of important parameters that are used in the assessment of cardiovascular function. An essential part of this analysis is the identification of the correct CMR views and quality control (QC) to detect artefacts that could affect the flow quantification. We propose a novel deep learning based framework for the fully-automated analysis of flow from full CMR scans that first carries out these view selection and QC steps using two sequential convolutional neural networks, followed by automatic aorta and pulmonary artery segmentation to enable the quantification of key flow parameters. Accuracy values of 0.958 and 0.914 were obtained for view classification and QC, respectively. For segmentation, Dice scores were $>$0.969 and the Bland-Altman plots indicated excellent agreement between manual and automatic peak flow values. In addition, we tested our pipeline on an external validation data set, with results indicating good robustness of the pipeline. This work was carried out using multivendor clinical data consisting of 986 cases, indicating the potential for the use of this pipeline in a clinical setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge