Julia A. Schnabel
on behalf of the PREDICTOM consortium
A Master Class on Reproducibility: A Student Hackathon on Advanced MRI Reconstruction Methods
Jan 26, 2026Abstract:We report the design, protocol, and outcomes of a student reproducibility hackathon focused on replicating the results of three influential MRI reconstruction papers: (a) MoDL, an unrolled model-based network with learned denoising; (b) HUMUS-Net, a hybrid unrolled multiscale CNN+Transformer architecture; and (c) an untrained, physics-regularized dynamic MRI method that uses a quantitative MR model for early stopping. We describe the setup of the hackathon and present reproduction outcomes alongside additional experiments, and we detail fundamental practices for building reproducible codebases.
Measuring and Aligning Abstraction in Vision-Language Models with Medical Taxonomies
Jan 21, 2026Abstract:Vision-Language Models show strong zero-shot performance for chest X-ray classification, but standard flat metrics fail to distinguish between clinically minor and severe errors. This work investigates how to quantify and mitigate abstraction errors by leveraging medical taxonomies. We benchmark several state-of-the-art VLMs using hierarchical metrics and introduce Catastrophic Abstraction Errors to capture cross-branch mistakes. Our results reveal substantial misalignment of VLMs with clinical taxonomies despite high flat performance. To address this, we propose risk-constrained thresholding and taxonomy-aware fine-tuning with radial embeddings, which reduce severe abstraction errors to below 2 per cent while maintaining competitive performance. These findings highlight the importance of hierarchical evaluation and representation-level alignment for safer and more clinically meaningful deployment of VLMs.
LocBAM: Advancing 3D Patch-Based Image Segmentation by Integrating Location Contex
Jan 21, 2026Abstract:Patch-based methods are widely used in 3D medical image segmentation to address memory constraints in processing high-resolution volumetric data. However, these approaches often neglect the patch's location within the global volume, which can limit segmentation performance when anatomical context is important. In this paper, we investigate the role of location context in patch-based 3D segmentation and propose a novel attention mechanism, LocBAM, that explicitly processes spatial information. Experiments on BTCV, AMOS22, and KiTS23 demonstrate that incorporating location context stabilizes training and improves segmentation performance, particularly under low patch-to-volume coverage where global context is missing. Furthermore, LocBAM consistently outperforms classical coordinate encoding via CoordConv. Code is publicly available at https://github.com/compai-lab/2026-ISBI-hooft
TomoGraphView: 3D Medical Image Classification with Omnidirectional Slice Representations and Graph Neural Networks
Nov 12, 2025Abstract:The growing number of medical tomography examinations has necessitated the development of automated methods capable of extracting comprehensive imaging features to facilitate downstream tasks such as tumor characterization, while assisting physicians in managing their growing workload. However, 3D medical image classification remains a challenging task due to the complex spatial relationships and long-range dependencies inherent in volumetric data. Training models from scratch suffers from low data regimes, and the absence of 3D large-scale multimodal datasets has limited the development of 3D medical imaging foundation models. Recent studies, however, have highlighted the potential of 2D vision foundation models, originally trained on natural images, as powerful feature extractors for medical image analysis. Despite these advances, existing approaches that apply 2D models to 3D volumes via slice-based decomposition remain suboptimal. Conventional volume slicing strategies, which rely on canonical planes such as axial, sagittal, or coronal, may inadequately capture the spatial extent of target structures when these are misaligned with standardized viewing planes. Furthermore, existing slice-wise aggregation strategies rarely account for preserving the volumetric structure, resulting in a loss of spatial coherence across slices. To overcome these limitations, we propose TomoGraphView, a novel framework that integrates omnidirectional volume slicing with spherical graph-based feature aggregation. We publicly share our accessible code base at http://github.com/compai-lab/2025-MedIA-kiechle and provide a user-friendly library for omnidirectional volume slicing at https://pypi.org/project/OmniSlicer.
Covariance Descriptors Meet General Vision Encoders: Riemannian Deep Learning for Medical Image Classification
Nov 06, 2025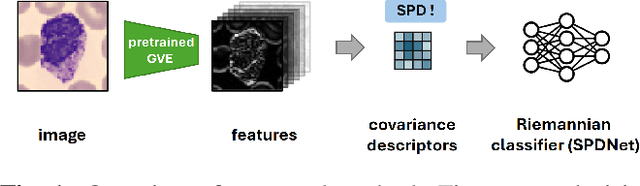
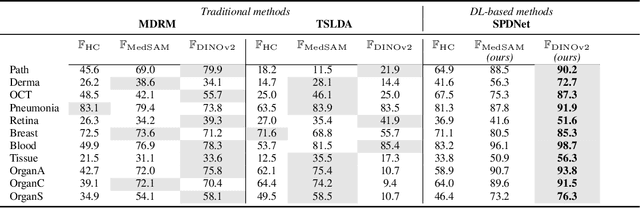
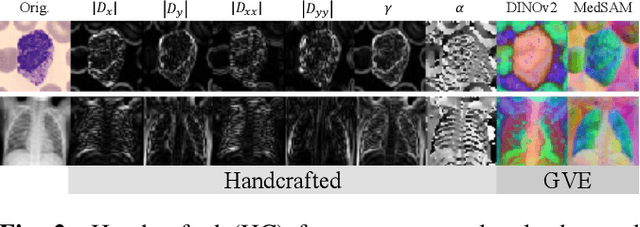
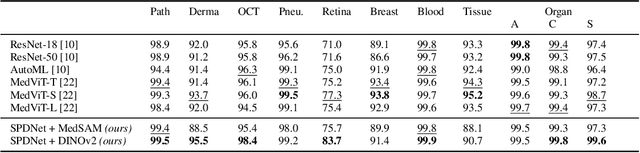
Abstract:Covariance descriptors capture second-order statistics of image features. They have shown strong performance in general computer vision tasks, but remain underexplored in medical imaging. We investigate their effectiveness for both conventional and learning-based medical image classification, with a particular focus on SPDNet, a classification network specifically designed for symmetric positive definite (SPD) matrices. We propose constructing covariance descriptors from features extracted by pre-trained general vision encoders (GVEs) and comparing them with handcrafted descriptors. Two GVEs - DINOv2 and MedSAM - are evaluated across eleven binary and multi-class datasets from the MedMNSIT benchmark. Our results show that covariance descriptors derived from GVE features consistently outperform those derived from handcrafted features. Moreover, SPDNet yields superior performance to state-of-the-art methods when combined with DINOv2 features. Our findings highlight the potential of combining covariance descriptors with powerful pretrained vision encoders for medical image analysis.
Learning to reason about rare diseases through retrieval-augmented agents
Nov 06, 2025Abstract:Rare diseases represent the long tail of medical imaging, where AI models often fail due to the scarcity of representative training data. In clinical workflows, radiologists frequently consult case reports and literature when confronted with unfamiliar findings. Following this line of reasoning, we introduce RADAR, Retrieval Augmented Diagnostic Reasoning Agents, an agentic system for rare disease detection in brain MRI. Our approach uses AI agents with access to external medical knowledge by embedding both case reports and literature using sentence transformers and indexing them with FAISS to enable efficient similarity search. The agent retrieves clinically relevant evidence to guide diagnostic decision making on unseen diseases, without the need of additional training. Designed as a model-agnostic reasoning module, RADAR can be seamlessly integrated with diverse large language models, consistently improving their rare pathology recognition and interpretability. On the NOVA dataset comprising 280 distinct rare diseases, RADAR achieves up to a 10.2% performance gain, with the strongest improvements observed for open source models such as DeepSeek. Beyond accuracy, the retrieved examples provide interpretable, literature grounded explanations, highlighting retrieval-augmented reasoning as a powerful paradigm for low-prevalence conditions in medical imaging.
INR meets Multi-Contrast MRI Reconstruction
Sep 05, 2025Abstract:Multi-contrast MRI sequences allow for the acquisition of images with varying tissue contrast within a single scan. The resulting multi-contrast images can be used to extract quantitative information on tissue microstructure. To make such multi-contrast sequences feasible for clinical routine, the usually very long scan times need to be shortened e.g. through undersampling in k-space. However, this comes with challenges for the reconstruction. In general, advanced reconstruction techniques such as compressed sensing or deep learning-based approaches can enable the acquisition of high-quality images despite the acceleration. In this work, we leverage redundant anatomical information of multi-contrast sequences to achieve even higher acceleration rates. We use undersampling patterns that capture the contrast information located at the k-space center, while performing complementary undersampling across contrasts for high frequencies. To reconstruct this highly sparse k-space data, we propose an implicit neural representation (INR) network that is ideal for using the complementary information acquired across contrasts as it jointly reconstructs all contrast images. We demonstrate the benefits of our proposed INR method by applying it to multi-contrast MRI using the MPnRAGE sequence, where it outperforms the state-of-the-art parallel imaging compressed sensing (PICS) reconstruction method, even at higher acceleration factors.
Knowledge to Sight: Reasoning over Visual Attributes via Knowledge Decomposition for Abnormality Grounding
Aug 06, 2025Abstract:In this work, we address the problem of grounding abnormalities in medical images, where the goal is to localize clinical findings based on textual descriptions. While generalist Vision-Language Models (VLMs) excel in natural grounding tasks, they often struggle in the medical domain due to rare, compositional, and domain-specific terms that are poorly aligned with visual patterns. Specialized medical VLMs address this challenge via large-scale domain pretraining, but at the cost of substantial annotation and computational resources. To overcome these limitations, we propose \textbf{Knowledge to Sight (K2Sight)}, a framework that introduces structured semantic supervision by decomposing clinical concepts into interpretable visual attributes, such as shape, density, and anatomical location. These attributes are distilled from domain ontologies and encoded into concise instruction-style prompts, which guide region-text alignment during training. Unlike conventional report-level supervision, our approach explicitly bridges domain knowledge and spatial structure, enabling data-efficient training of compact models. We train compact models with 0.23B and 2B parameters using only 1.5\% of the data required by state-of-the-art medical VLMs. Despite their small size and limited training data, these models achieve performance on par with or better than 7B+ medical VLMs, with up to 9.82\% improvement in $mAP_{50}$. Code and models: \href{https://lijunrio.github.io/K2Sight/}{\textcolor{SOTAPink}{https://lijunrio.github.io/K2Sight/}}.
NOVA: A Benchmark for Anomaly Localization and Clinical Reasoning in Brain MRI
May 20, 2025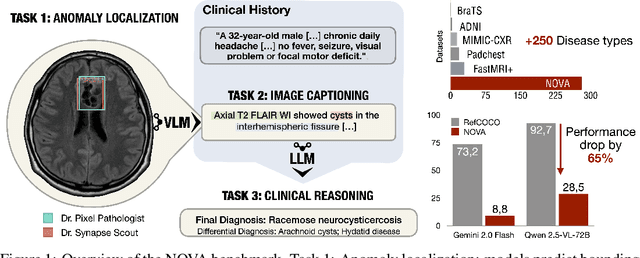

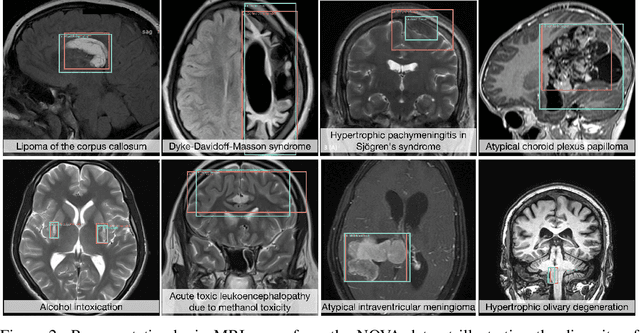
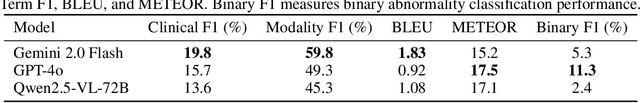
Abstract:In many real-world applications, deployed models encounter inputs that differ from the data seen during training. Out-of-distribution detection identifies whether an input stems from an unseen distribution, while open-world recognition flags such inputs to ensure the system remains robust as ever-emerging, previously $unknown$ categories appear and must be addressed without retraining. Foundation and vision-language models are pre-trained on large and diverse datasets with the expectation of broad generalization across domains, including medical imaging. However, benchmarking these models on test sets with only a few common outlier types silently collapses the evaluation back to a closed-set problem, masking failures on rare or truly novel conditions encountered in clinical use. We therefore present $NOVA$, a challenging, real-life $evaluation-only$ benchmark of $\sim$900 brain MRI scans that span 281 rare pathologies and heterogeneous acquisition protocols. Each case includes rich clinical narratives and double-blinded expert bounding-box annotations. Together, these enable joint assessment of anomaly localisation, visual captioning, and diagnostic reasoning. Because NOVA is never used for training, it serves as an $extreme$ stress-test of out-of-distribution generalisation: models must bridge a distribution gap both in sample appearance and in semantic space. Baseline results with leading vision-language models (GPT-4o, Gemini 2.0 Flash, and Qwen2.5-VL-72B) reveal substantial performance drops across all tasks, establishing NOVA as a rigorous testbed for advancing models that can detect, localize, and reason about truly unknown anomalies.
Semantic Alignment of Unimodal Medical Text and Vision Representations
Mar 06, 2025Abstract:General-purpose AI models, particularly those designed for text and vision, demonstrate impressive versatility across a wide range of deep-learning tasks. However, they often underperform in specialised domains like medical imaging, where domain-specific solutions or alternative knowledge transfer approaches are typically required. Recent studies have noted that general-purpose models can exhibit similar latent spaces when processing semantically related data, although this alignment does not occur naturally. Building on this insight, it has been shown that applying a simple transformation - at most affine - estimated from a subset of semantically corresponding samples, known as anchors, enables model stitching across diverse training paradigms, architectures, and modalities. In this paper, we explore how semantic alignment - estimating transformations between anchors - can bridge general-purpose AI with specialised medical knowledge. Using multiple public chest X-ray datasets, we demonstrate that model stitching across model architectures allows general models to integrate domain-specific knowledge without additional training, leading to improved performance on medical tasks. Furthermore, we introduce a novel zero-shot classification approach for unimodal vision encoders that leverages semantic alignment across modalities. Our results show that our method not only outperforms general multimodal models but also approaches the performance levels of fully trained, medical-specific multimodal solutions
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge