Lina Felsner
A Master Class on Reproducibility: A Student Hackathon on Advanced MRI Reconstruction Methods
Jan 26, 2026Abstract:We report the design, protocol, and outcomes of a student reproducibility hackathon focused on replicating the results of three influential MRI reconstruction papers: (a) MoDL, an unrolled model-based network with learned denoising; (b) HUMUS-Net, a hybrid unrolled multiscale CNN+Transformer architecture; and (c) an untrained, physics-regularized dynamic MRI method that uses a quantitative MR model for early stopping. We describe the setup of the hackathon and present reproduction outcomes alongside additional experiments, and we detail fundamental practices for building reproducible codebases.
Uncertainty-guided Generation of Dark-field Radiographs
Jan 22, 2026Abstract:X-ray dark-field radiography provides complementary diagnostic information to conventional attenuation imaging by visualizing microstructural tissue changes through small-angle scattering. However, the limited availability of such data poses challenges for developing robust deep learning models. In this work, we present the first framework for generating dark-field images directly from standard attenuation chest X-rays using an Uncertainty-Guided Progressive Generative Adversarial Network. The model incorporates both aleatoric and epistemic uncertainty to improve interpretability and reliability. Experiments demonstrate high structural fidelity of the generated images, with consistent improvement of quantitative metrics across stages. Furthermore, out-of-distribution evaluation confirms that the proposed model generalizes well. Our results indicate that uncertainty-guided generative modeling enables realistic dark-field image synthesis and provides a reliable foundation for future clinical applications.
TomoGraphView: 3D Medical Image Classification with Omnidirectional Slice Representations and Graph Neural Networks
Nov 12, 2025Abstract:The growing number of medical tomography examinations has necessitated the development of automated methods capable of extracting comprehensive imaging features to facilitate downstream tasks such as tumor characterization, while assisting physicians in managing their growing workload. However, 3D medical image classification remains a challenging task due to the complex spatial relationships and long-range dependencies inherent in volumetric data. Training models from scratch suffers from low data regimes, and the absence of 3D large-scale multimodal datasets has limited the development of 3D medical imaging foundation models. Recent studies, however, have highlighted the potential of 2D vision foundation models, originally trained on natural images, as powerful feature extractors for medical image analysis. Despite these advances, existing approaches that apply 2D models to 3D volumes via slice-based decomposition remain suboptimal. Conventional volume slicing strategies, which rely on canonical planes such as axial, sagittal, or coronal, may inadequately capture the spatial extent of target structures when these are misaligned with standardized viewing planes. Furthermore, existing slice-wise aggregation strategies rarely account for preserving the volumetric structure, resulting in a loss of spatial coherence across slices. To overcome these limitations, we propose TomoGraphView, a novel framework that integrates omnidirectional volume slicing with spherical graph-based feature aggregation. We publicly share our accessible code base at http://github.com/compai-lab/2025-MedIA-kiechle and provide a user-friendly library for omnidirectional volume slicing at https://pypi.org/project/OmniSlicer.
Motion-Robust T2* Quantification from Gradient Echo MRI with Physics-Informed Deep Learning
Feb 24, 2025Abstract:Purpose: T2* quantification from gradient echo magnetic resonance imaging is particularly affected by subject motion due to the high sensitivity to magnetic field inhomogeneities, which are influenced by motion and might cause signal loss. Thus, motion correction is crucial to obtain high-quality T2* maps. Methods: We extend our previously introduced learning-based physics-informed motion correction method, PHIMO, by utilizing acquisition knowledge to enhance the reconstruction performance for challenging motion patterns and increase PHIMO's robustness to varying strengths of magnetic field inhomogeneities across the brain. We perform comprehensive evaluations regarding motion detection accuracy and image quality for data with simulated and real motion. Results: Our extended version of PHIMO outperforms the learning-based baseline methods both qualitatively and quantitatively with respect to line detection and image quality. Moreover, PHIMO performs on-par with a conventional state-of-the-art motion correction method for T2* quantification from gradient echo MRI, which relies on redundant data acquisition. Conclusion: PHIMO's competitive motion correction performance, combined with a reduction in acquisition time by over 40% compared to the state-of-the-art method, make it a promising solution for motion-robust T2* quantification in research settings and clinical routine.
Graph Neural Networks: A suitable Alternative to MLPs in Latent 3D Medical Image Classification?
Jul 24, 2024



Abstract:Recent studies have underscored the capabilities of natural imaging foundation models to serve as powerful feature extractors, even in a zero-shot setting for medical imaging data. Most commonly, a shallow multi-layer perceptron (MLP) is appended to the feature extractor to facilitate end-to-end learning and downstream prediction tasks such as classification, thus representing the de facto standard. However, as graph neural networks (GNNs) have become a practicable choice for various tasks in medical research in the recent past, we direct attention to the question of how effective GNNs are compared to MLP prediction heads for the task of 3D medical image classification, proposing them as a potential alternative. In our experiments, we devise a subject-level graph for each volumetric dataset instance. Therein latent representations of all slices in the volume, encoded through a DINOv2 pretrained vision transformer (ViT), constitute the nodes and their respective node features. We use public datasets to compare the classification heads numerically and evaluate various graph construction and graph convolution methods in our experiments. Our findings show enhancements of the GNN in classification performance and substantial improvements in runtime compared to an MLP prediction head. Additional robustness evaluations further validate the promising performance of the GNN, promoting them as a suitable alternative to traditional MLP classification heads. Our code is publicly available at: https://github.com/compai-lab/2024-miccai-grail-kiechle
Progressive Growing of Patch Size: Resource-Efficient Curriculum Learning for Dense Prediction Tasks
Jul 11, 2024Abstract:In this work, we introduce Progressive Growing of Patch Size, a resource-efficient implicit curriculum learning approach for dense prediction tasks. Our curriculum approach is defined by growing the patch size during model training, which gradually increases the task's difficulty. We integrated our curriculum into the nnU-Net framework and evaluated the methodology on all 10 tasks of the Medical Segmentation Decathlon. With our approach, we are able to substantially reduce runtime, computational costs, and CO2 emissions of network training compared to classical constant patch size training. In our experiments, the curriculum approach resulted in improved convergence. We are able to outperform standard nnU-Net training, which is trained with constant patch size, in terms of Dice Score on 7 out of 10 MSD tasks while only spending roughly 50% of the original training runtime. To the best of our knowledge, our Progressive Growing of Patch Size is the first successful employment of a sample-length curriculum in the form of patch size in the field of computer vision. Our code is publicly available at https://github.com/compai-lab/2024-miccai-fischer.
Data-Driven Tissue- and Subject-Specific Elastic Regularization for Medical Image Registration
Jul 05, 2024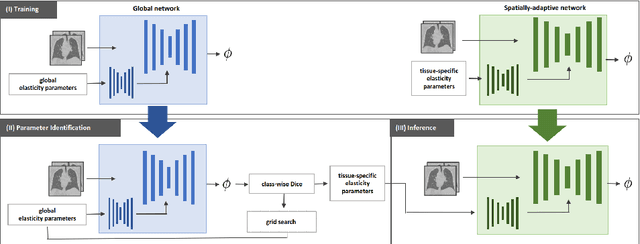
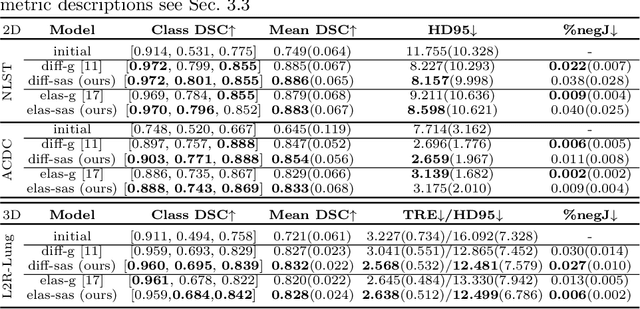
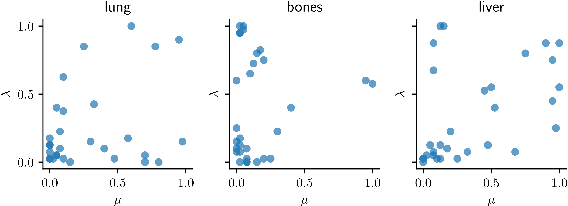
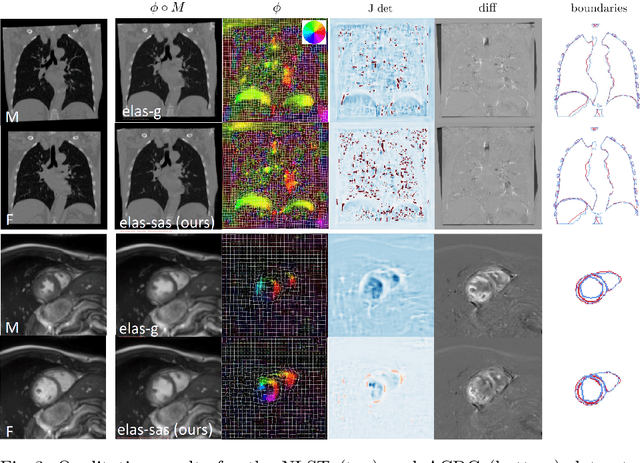
Abstract:Physics-inspired regularization is desired for intra-patient image registration since it can effectively capture the biomechanical characteristics of anatomical structures. However, a major challenge lies in the reliance on physical parameters: Parameter estimations vary widely across the literature, and the physical properties themselves are inherently subject-specific. In this work, we introduce a novel data-driven method that leverages hypernetworks to learn the tissue-dependent elasticity parameters of an elastic regularizer. Notably, our approach facilitates the estimation of patient-specific parameters without the need to retrain the network. We evaluate our method on three publicly available 2D and 3D lung CT and cardiac MR datasets. We find that with our proposed subject-specific tissue-dependent regularization, a higher registration quality is achieved across all datasets compared to using a global regularizer. The code is available at https://github.com/compai-lab/2024-miccai-reithmeir.
Multi-Image Visual Question Answering for Unsupervised Anomaly Detection
Apr 11, 2024



Abstract:Unsupervised anomaly detection enables the identification of potential pathological areas by juxtaposing original images with their pseudo-healthy reconstructions generated by models trained exclusively on normal images. However, the clinical interpretation of resultant anomaly maps presents a challenge due to a lack of detailed, understandable explanations. Recent advancements in language models have shown the capability of mimicking human-like understanding and providing detailed descriptions. This raises an interesting question: \textit{How can language models be employed to make the anomaly maps more explainable?} To the best of our knowledge, we are the first to leverage a language model for unsupervised anomaly detection, for which we construct a dataset with different questions and answers. Additionally, we present a novel multi-image visual question answering framework tailored for anomaly detection, incorporating diverse feature fusion strategies to enhance visual knowledge extraction. Our experiments reveal that the framework, augmented by our new Knowledge Q-Former module, adeptly answers questions on the anomaly detection dataset. Besides, integrating anomaly maps as inputs distinctly aids in improving the detection of unseen pathologies.
Learned Cone-Beam CT Reconstruction Using Neural Ordinary Differential Equations
Jan 19, 2022
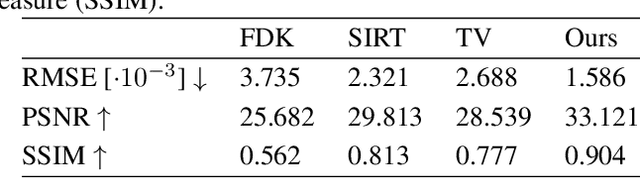

Abstract:Learned iterative reconstruction algorithms for inverse problems offer the flexibility to combine analytical knowledge about the problem with modules learned from data. This way, they achieve high reconstruction performance while ensuring consistency with the measured data. In computed tomography, extending such approaches from 2D fan-beam to 3D cone-beam data is challenging due to the prohibitively high GPU memory that would be needed to train such models. This paper proposes to use neural ordinary differential equations to solve the reconstruction problem in a residual formulation via numerical integration. For training, there is no need to backpropagate through several unrolled network blocks nor through the internals of the solver. Instead, the gradients are obtained very memory-efficiently in the neural ODE setting allowing for training on a single consumer graphics card. The method is able to reduce the root mean squared error by over 30% compared to the best performing classical iterative reconstruction algorithm and produces high quality cone-beam reconstructions even in a sparse view scenario.
X-ray Scatter Estimation Using Deep Splines
Jan 22, 2021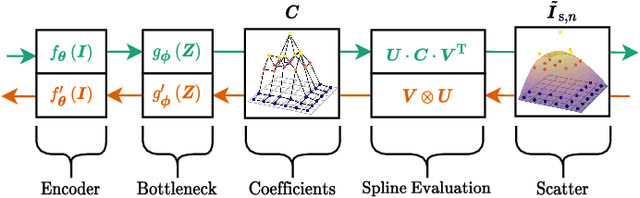
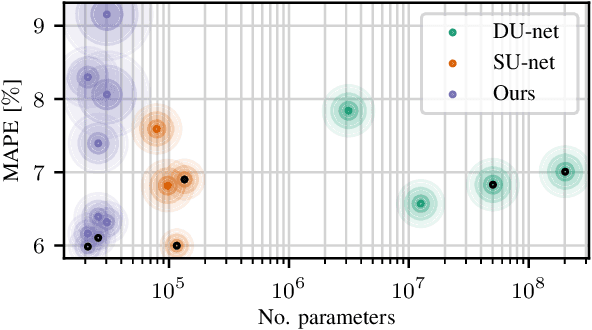
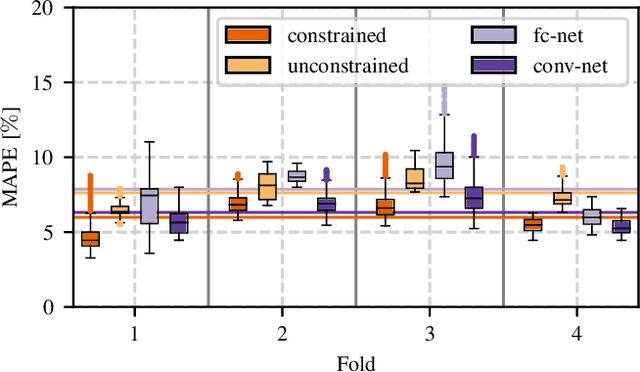
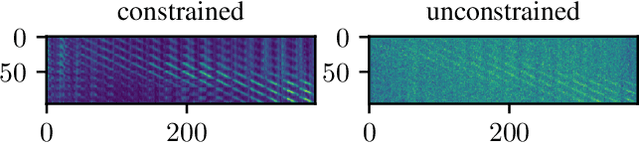
Abstract:Algorithmic X-ray scatter compensation is a desirable technique in flat-panel X-ray imaging and cone-beam computed tomography. State-of-the-art U-net based image translation approaches yielded promising results. As there are no physics constraints applied to the output of the U-Net, it cannot be ruled out that it yields spurious results. Unfortunately, those may be misleading in the context of medical imaging. To overcome this problem, we propose to embed B-splines as a known operator into neural networks. This inherently limits their predictions to well-behaved and smooth functions. In a study using synthetic head and thorax data as well as real thorax phantom data, we found that our approach performed on par with U-net when comparing both algorithms based on quantitative performance metrics. However, our approach not only reduces runtime and parameter complexity, but we also found it much more robust to unseen noise levels. While the U-net responded with visible artifacts, our approach preserved the X-ray signal's frequency characteristics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge