Johannes Kiechle
TomoGraphView: 3D Medical Image Classification with Omnidirectional Slice Representations and Graph Neural Networks
Nov 12, 2025Abstract:The growing number of medical tomography examinations has necessitated the development of automated methods capable of extracting comprehensive imaging features to facilitate downstream tasks such as tumor characterization, while assisting physicians in managing their growing workload. However, 3D medical image classification remains a challenging task due to the complex spatial relationships and long-range dependencies inherent in volumetric data. Training models from scratch suffers from low data regimes, and the absence of 3D large-scale multimodal datasets has limited the development of 3D medical imaging foundation models. Recent studies, however, have highlighted the potential of 2D vision foundation models, originally trained on natural images, as powerful feature extractors for medical image analysis. Despite these advances, existing approaches that apply 2D models to 3D volumes via slice-based decomposition remain suboptimal. Conventional volume slicing strategies, which rely on canonical planes such as axial, sagittal, or coronal, may inadequately capture the spatial extent of target structures when these are misaligned with standardized viewing planes. Furthermore, existing slice-wise aggregation strategies rarely account for preserving the volumetric structure, resulting in a loss of spatial coherence across slices. To overcome these limitations, we propose TomoGraphView, a novel framework that integrates omnidirectional volume slicing with spherical graph-based feature aggregation. We publicly share our accessible code base at http://github.com/compai-lab/2025-MedIA-kiechle and provide a user-friendly library for omnidirectional volume slicing at https://pypi.org/project/OmniSlicer.
Graph Neural Networks: A suitable Alternative to MLPs in Latent 3D Medical Image Classification?
Jul 24, 2024



Abstract:Recent studies have underscored the capabilities of natural imaging foundation models to serve as powerful feature extractors, even in a zero-shot setting for medical imaging data. Most commonly, a shallow multi-layer perceptron (MLP) is appended to the feature extractor to facilitate end-to-end learning and downstream prediction tasks such as classification, thus representing the de facto standard. However, as graph neural networks (GNNs) have become a practicable choice for various tasks in medical research in the recent past, we direct attention to the question of how effective GNNs are compared to MLP prediction heads for the task of 3D medical image classification, proposing them as a potential alternative. In our experiments, we devise a subject-level graph for each volumetric dataset instance. Therein latent representations of all slices in the volume, encoded through a DINOv2 pretrained vision transformer (ViT), constitute the nodes and their respective node features. We use public datasets to compare the classification heads numerically and evaluate various graph construction and graph convolution methods in our experiments. Our findings show enhancements of the GNN in classification performance and substantial improvements in runtime compared to an MLP prediction head. Additional robustness evaluations further validate the promising performance of the GNN, promoting them as a suitable alternative to traditional MLP classification heads. Our code is publicly available at: https://github.com/compai-lab/2024-miccai-grail-kiechle
Progressive Growing of Patch Size: Resource-Efficient Curriculum Learning for Dense Prediction Tasks
Jul 11, 2024Abstract:In this work, we introduce Progressive Growing of Patch Size, a resource-efficient implicit curriculum learning approach for dense prediction tasks. Our curriculum approach is defined by growing the patch size during model training, which gradually increases the task's difficulty. We integrated our curriculum into the nnU-Net framework and evaluated the methodology on all 10 tasks of the Medical Segmentation Decathlon. With our approach, we are able to substantially reduce runtime, computational costs, and CO2 emissions of network training compared to classical constant patch size training. In our experiments, the curriculum approach resulted in improved convergence. We are able to outperform standard nnU-Net training, which is trained with constant patch size, in terms of Dice Score on 7 out of 10 MSD tasks while only spending roughly 50% of the original training runtime. To the best of our knowledge, our Progressive Growing of Patch Size is the first successful employment of a sample-length curriculum in the form of patch size in the field of computer vision. Our code is publicly available at https://github.com/compai-lab/2024-miccai-fischer.
Mask the Unknown: Assessing Different Strategies to Handle Weak Annotations in the MICCAI2023 Mediastinal Lymph Node Quantification Challenge
Jun 20, 2024
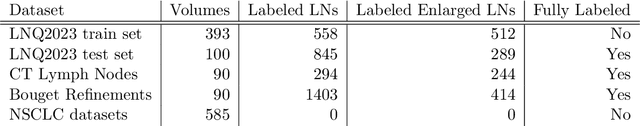
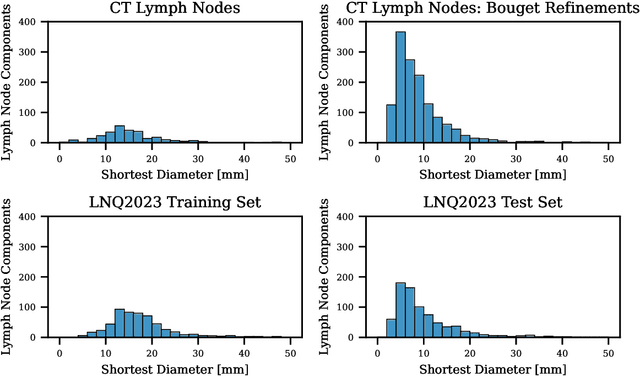
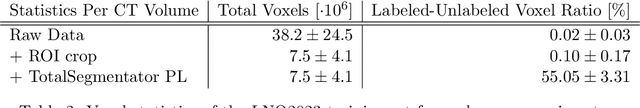
Abstract:Pathological lymph node delineation is crucial in cancer diagnosis, progression assessment, and treatment planning. The MICCAI 2023 Lymph Node Quantification Challenge published the first public dataset for pathological lymph node segmentation in the mediastinum. As lymph node annotations are expensive, the challenge was formed as a weakly supervised learning task, where only a subset of all lymph nodes in the training set have been annotated. For the challenge submission, multiple methods for training on these weakly supervised data were explored, including noisy label training, loss masking of unlabeled data, and an approach that integrated the TotalSegmentator toolbox as a form of pseudo labeling in order to reduce the number of unknown voxels. Furthermore, multiple public TCIA datasets were incorporated into the training to improve the performance of the deep learning model. Our submitted model achieved a Dice score of 0.628 and an average symmetric surface distance of 5.8~mm on the challenge test set. With our submitted model, we accomplished third rank in the MICCAI2023 LNQ challenge. A finding of our analysis was that the integration of all visible, including non-pathological, lymph nodes improved the overall segmentation performance on pathological lymph nodes of the test set. Furthermore, segmentation models trained only on clinically enlarged lymph nodes, as given in the challenge scenario, could not generalize to smaller pathological lymph nodes. The code and model for the challenge submission are available at \url{https://gitlab.lrz.de/compai/MediastinalLymphNodeSegmentation}.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:008
Disentangling Hippocampal Shape Variations: A Study of Neurological Disorders Using Graph Variational Autoencoder with Contrastive Learning
Mar 31, 2024



Abstract:This paper presents a comprehensive study focused on disentangling hippocampal shape variations from diffusion tensor imaging (DTI) datasets within the context of neurological disorders. Leveraging a Graph Variational Autoencoder (VAE) enhanced with Supervised Contrastive Learning, our approach aims to improve interpretability by disentangling two distinct latent variables corresponding to age and the presence of diseases. In our ablation study, we investigate a range of VAE architectures and contrastive loss functions, showcasing the enhanced disentanglement capabilities of our approach. This evaluation uses synthetic 3D torus mesh data and real 3D hippocampal mesh datasets derived from the DTI hippocampal dataset. Our supervised disentanglement model outperforms several state-of-the-art (SOTA) methods like attribute and guided VAEs in terms of disentanglement scores. Our model distinguishes between age groups and disease status in patients with Multiple Sclerosis (MS) using the hippocampus data. Our Graph VAE with Supervised Contrastive Learning shows the volume changes of the hippocampus of MS populations at different ages, and the result is consistent with the current neuroimaging literature. This research provides valuable insights into the relationship between neurological disorder and hippocampal shape changes in different age groups of MS populations using a Graph VAE with Supervised Contrastive loss.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge