Hannah Eichhorn
on behalf of the PREDICTOM consortium
A Master Class on Reproducibility: A Student Hackathon on Advanced MRI Reconstruction Methods
Jan 26, 2026Abstract:We report the design, protocol, and outcomes of a student reproducibility hackathon focused on replicating the results of three influential MRI reconstruction papers: (a) MoDL, an unrolled model-based network with learned denoising; (b) HUMUS-Net, a hybrid unrolled multiscale CNN+Transformer architecture; and (c) an untrained, physics-regularized dynamic MRI method that uses a quantitative MR model for early stopping. We describe the setup of the hackathon and present reproduction outcomes alongside additional experiments, and we detail fundamental practices for building reproducible codebases.
INR meets Multi-Contrast MRI Reconstruction
Sep 05, 2025Abstract:Multi-contrast MRI sequences allow for the acquisition of images with varying tissue contrast within a single scan. The resulting multi-contrast images can be used to extract quantitative information on tissue microstructure. To make such multi-contrast sequences feasible for clinical routine, the usually very long scan times need to be shortened e.g. through undersampling in k-space. However, this comes with challenges for the reconstruction. In general, advanced reconstruction techniques such as compressed sensing or deep learning-based approaches can enable the acquisition of high-quality images despite the acceleration. In this work, we leverage redundant anatomical information of multi-contrast sequences to achieve even higher acceleration rates. We use undersampling patterns that capture the contrast information located at the k-space center, while performing complementary undersampling across contrasts for high frequencies. To reconstruct this highly sparse k-space data, we propose an implicit neural representation (INR) network that is ideal for using the complementary information acquired across contrasts as it jointly reconstructs all contrast images. We demonstrate the benefits of our proposed INR method by applying it to multi-contrast MRI using the MPnRAGE sequence, where it outperforms the state-of-the-art parallel imaging compressed sensing (PICS) reconstruction method, even at higher acceleration factors.
Motion-Robust T2* Quantification from Gradient Echo MRI with Physics-Informed Deep Learning
Feb 24, 2025Abstract:Purpose: T2* quantification from gradient echo magnetic resonance imaging is particularly affected by subject motion due to the high sensitivity to magnetic field inhomogeneities, which are influenced by motion and might cause signal loss. Thus, motion correction is crucial to obtain high-quality T2* maps. Methods: We extend our previously introduced learning-based physics-informed motion correction method, PHIMO, by utilizing acquisition knowledge to enhance the reconstruction performance for challenging motion patterns and increase PHIMO's robustness to varying strengths of magnetic field inhomogeneities across the brain. We perform comprehensive evaluations regarding motion detection accuracy and image quality for data with simulated and real motion. Results: Our extended version of PHIMO outperforms the learning-based baseline methods both qualitatively and quantitatively with respect to line detection and image quality. Moreover, PHIMO performs on-par with a conventional state-of-the-art motion correction method for T2* quantification from gradient echo MRI, which relies on redundant data acquisition. Conclusion: PHIMO's competitive motion correction performance, combined with a reduction in acquisition time by over 40% compared to the state-of-the-art method, make it a promising solution for motion-robust T2* quantification in research settings and clinical routine.
PISCO: Self-Supervised k-Space Regularization for Improved Neural Implicit k-Space Representations of Dynamic MRI
Jan 16, 2025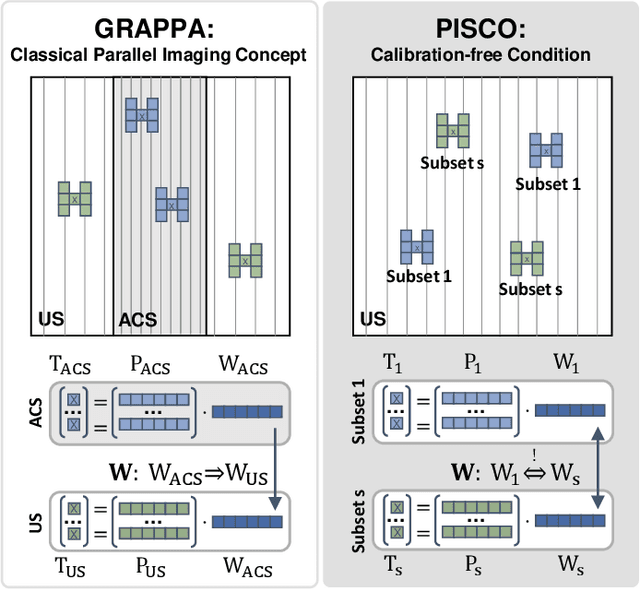
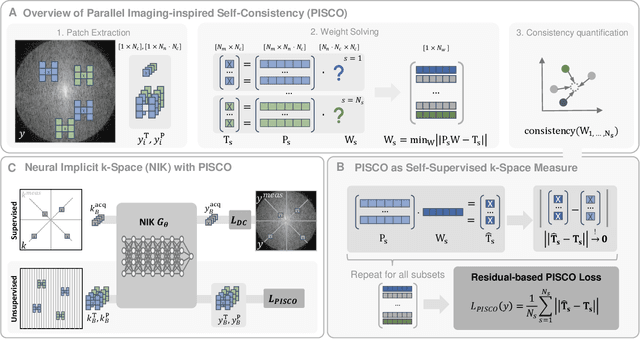
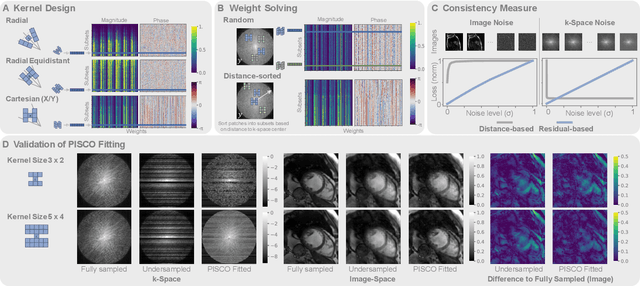
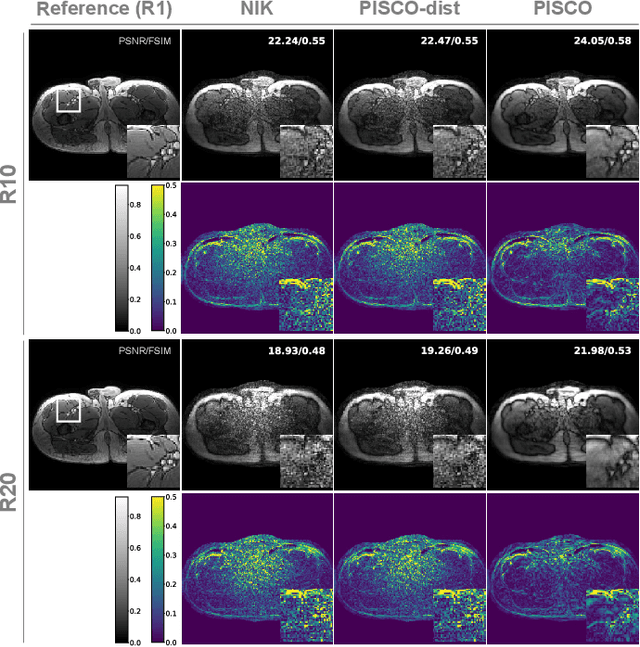
Abstract:Neural implicit k-space representations (NIK) have shown promising results for dynamic magnetic resonance imaging (MRI) at high temporal resolutions. Yet, reducing acquisition time, and thereby available training data, results in severe performance drops due to overfitting. To address this, we introduce a novel self-supervised k-space loss function $\mathcal{L}_\mathrm{PISCO}$, applicable for regularization of NIK-based reconstructions. The proposed loss function is based on the concept of parallel imaging-inspired self-consistency (PISCO), enforcing a consistent global k-space neighborhood relationship without requiring additional data. Quantitative and qualitative evaluations on static and dynamic MR reconstructions show that integrating PISCO significantly improves NIK representations. Particularly for high acceleration factors (R$\geq$54), NIK with PISCO achieves superior spatio-temporal reconstruction quality compared to state-of-the-art methods. Furthermore, an extensive analysis of the loss assumptions and stability shows PISCO's potential as versatile self-supervised k-space loss function for further applications and architectures. Code is available at: https://github.com/compai-lab/2025-pisco-spieker
Self-Supervised k-Space Regularization for Motion-Resolved Abdominal MRI Using Neural Implicit k-Space Representation
Apr 12, 2024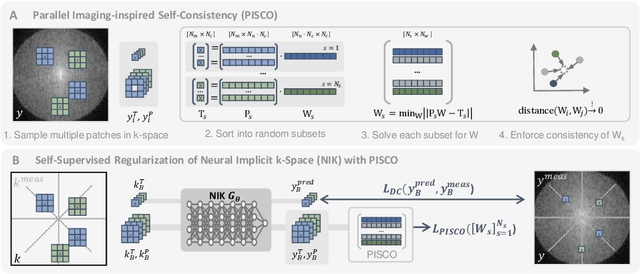
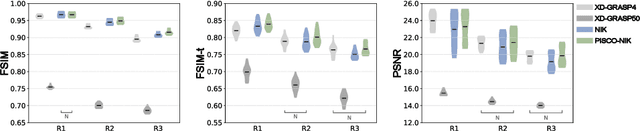
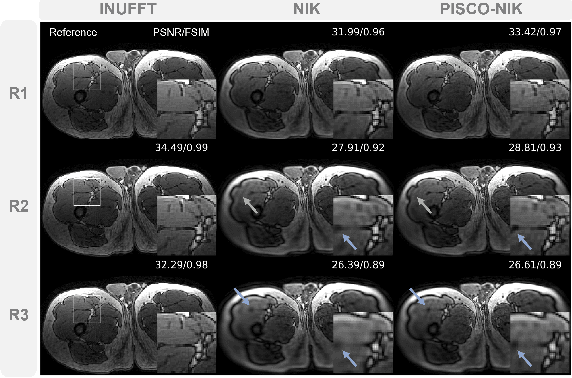
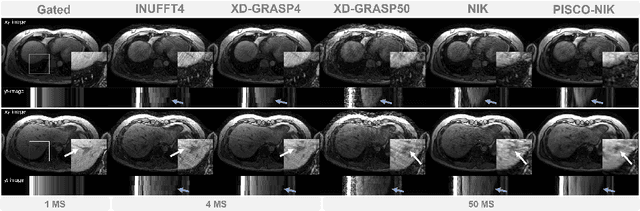
Abstract:Neural implicit k-space representations have shown promising results for dynamic MRI at high temporal resolutions. Yet, their exclusive training in k-space limits the application of common image regularization methods to improve the final reconstruction. In this work, we introduce the concept of parallel imaging-inspired self-consistency (PISCO), which we incorporate as novel self-supervised k-space regularization enforcing a consistent neighborhood relationship. At no additional data cost, the proposed regularization significantly improves neural implicit k-space reconstructions on simulated data. Abdominal in-vivo reconstructions using PISCO result in enhanced spatio-temporal image quality compared to state-of-the-art methods. Code is available at https://github.com/vjspi/PISCO-NIK.
Physics-Informed Deep Learning for Motion-Corrected Reconstruction of Quantitative Brain MRI
Mar 13, 2024Abstract:We propose PHIMO, a physics-informed learning-based motion correction method tailored to quantitative MRI. PHIMO leverages information from the signal evolution to exclude motion-corrupted k-space lines from a data-consistent reconstruction. We demonstrate the potential of PHIMO for the application of T2* quantification from gradient echo MRI, which is particularly sensitive to motion due to its sensitivity to magnetic field inhomogeneities. A state-of-the-art technique for motion correction requires redundant acquisition of the k-space center, prolonging the acquisition. We show that PHIMO can detect and exclude intra-scan motion events and, thus, correct for severe motion artifacts. PHIMO approaches the performance of the state-of-the-art motion correction method, while substantially reducing the acquisition time by over 40%, facilitating clinical applicability. Our code is available at https://github.com/HannahEichhorn/PHIMO.
ICoNIK: Generating Respiratory-Resolved Abdominal MR Reconstructions Using Neural Implicit Representations in k-Space
Aug 17, 2023


Abstract:Motion-resolved reconstruction for abdominal magnetic resonance imaging (MRI) remains a challenge due to the trade-off between residual motion blurring caused by discretized motion states and undersampling artefacts. In this work, we propose to generate blurring-free motion-resolved abdominal reconstructions by learning a neural implicit representation directly in k-space (NIK). Using measured sampling points and a data-derived respiratory navigator signal, we train a network to generate continuous signal values. To aid the regularization of sparsely sampled regions, we introduce an additional informed correction layer (ICo), which leverages information from neighboring regions to correct NIK's prediction. Our proposed generative reconstruction methods, NIK and ICoNIK, outperform standard motion-resolved reconstruction techniques and provide a promising solution to address motion artefacts in abdominal MRI.
Deep Learning for Retrospective Motion Correction in MRI: A Comprehensive Review
May 11, 2023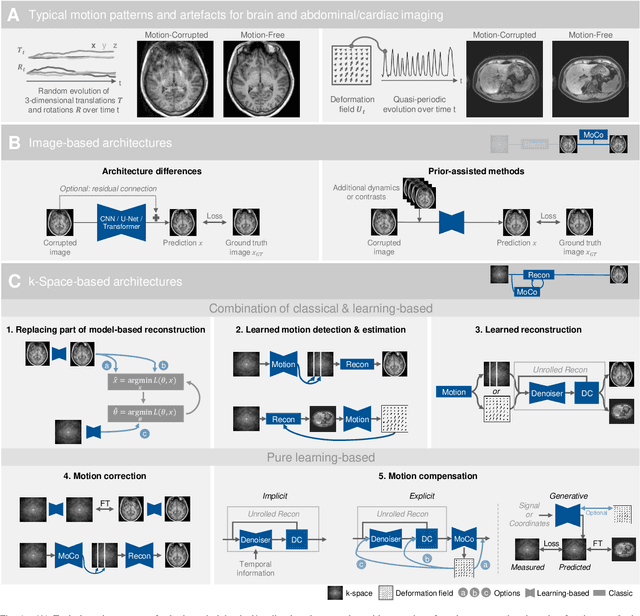
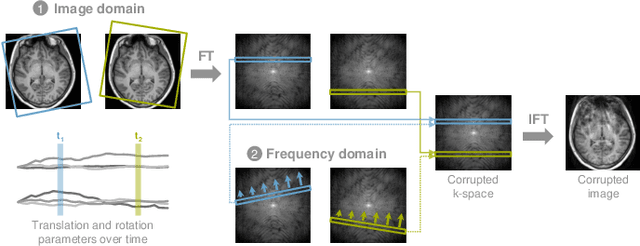
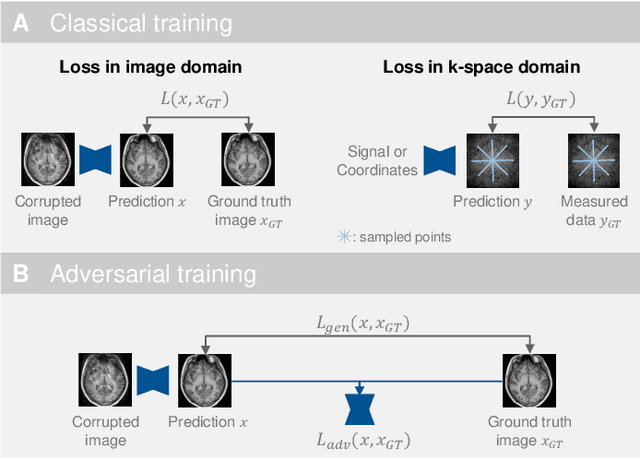
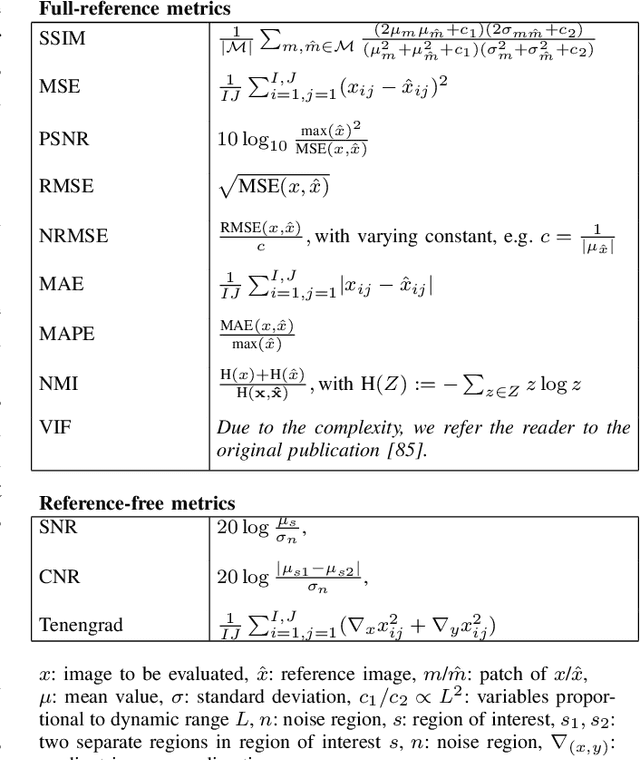
Abstract:Motion represents one of the major challenges in magnetic resonance imaging (MRI). Since the MR signal is acquired in frequency space, any motion of the imaged object leads to complex artefacts in the reconstructed image in addition to other MR imaging artefacts. Deep learning has been frequently proposed for motion correction at several stages of the reconstruction process. The wide range of MR acquisition sequences, anatomies and pathologies of interest, and motion patterns (rigid vs. deformable and random vs. regular) makes a comprehensive solution unlikely. To facilitate the transfer of ideas between different applications, this review provides a detailed overview of proposed methods for learning-based motion correction in MRI together with their common challenges and potentials. This review identifies differences and synergies in underlying data usage, architectures and evaluation strategies. We critically discuss general trends and outline future directions, with the aim to enhance interaction between different application areas and research fields.
Deep Learning-Based Detection of Motion-Affected k-Space Lines for T2*-Weighted MRI
Mar 20, 2023Abstract:T2*-weighted gradient echo MR imaging is strongly impacted by subject head motion due to motion-related changes in B0 inhomogeneities. Within the oxygenation-sensitive mqBOLD protocol, even mild motion during the acquisition of the T2*-weighted data propagates into errors in derived quantitative parameter maps. In order to correct these images without the need of repeated measurements, we propose to learn a classification of motion-affected k-space lines. To test this, we perform realistic motion simulations including motion-induced field inhomogeneity changes for supervised training. To detect the presence of motion in each phase encoding line, we train a convolutional neural network, leveraging the multi-echo information of the T2*-weighted images. The proposed network accurately detects motion-affected k-space lines for simulated displacements of $\geq$ 0.5mm (accuracy on test set: 92.5%). Finally, we show example reconstructions where we include these classification labels as weights in the data consistency term of an iterative reconstruction procedure, opening up exciting opportunities of k-space line detection in combination with more powerful reconstruction methods.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge