Veronika Cheplygina
Medical Imaging AI Competitions Lack Fairness
Dec 19, 2025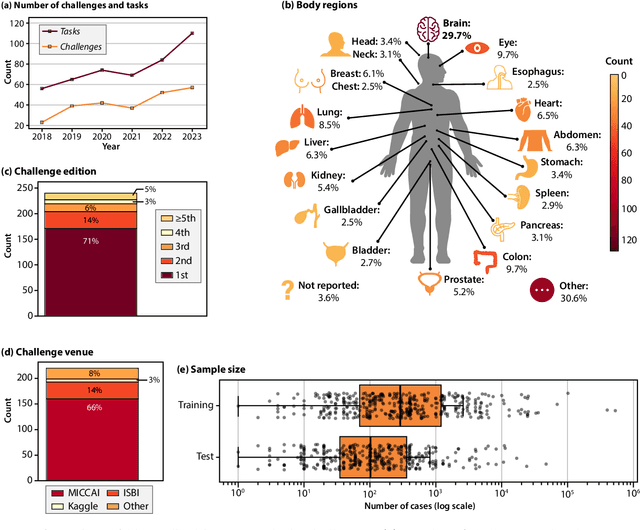
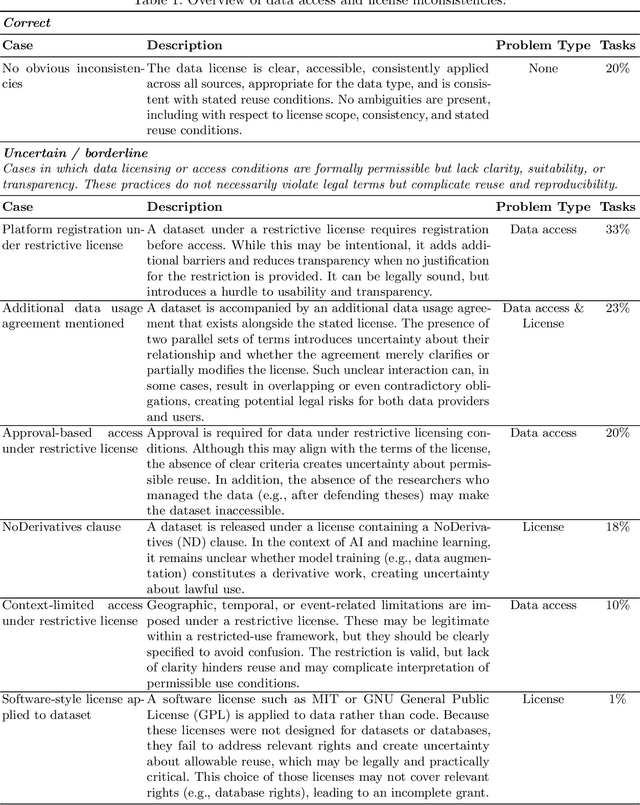
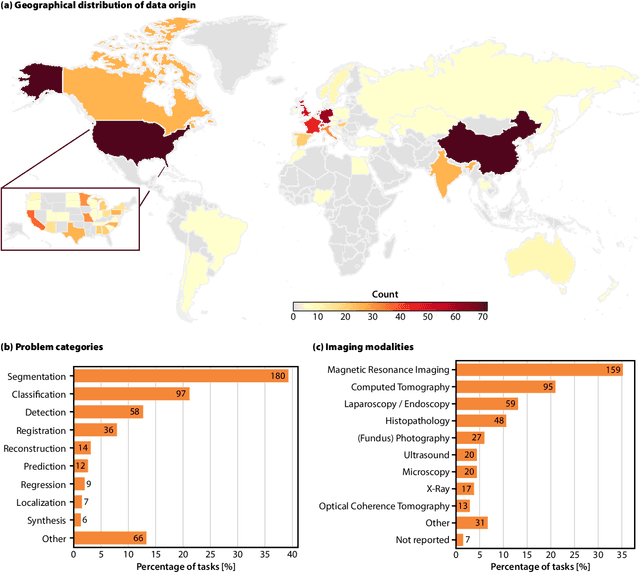
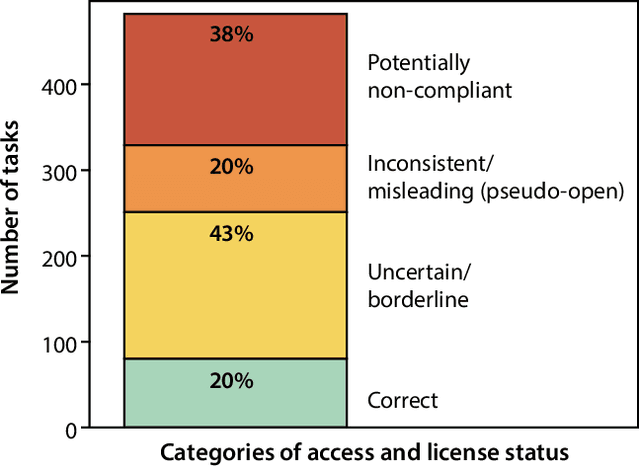
Abstract:Benchmarking competitions are central to the development of artificial intelligence (AI) in medical imaging, defining performance standards and shaping methodological progress. However, it remains unclear whether these benchmarks provide data that are sufficiently representative, accessible, and reusable to support clinically meaningful AI. In this work, we assess fairness along two complementary dimensions: (1) whether challenge datasets are representative of real-world clinical diversity, and (2) whether they are accessible and legally reusable in line with the FAIR principles. To address this question, we conducted a large-scale systematic study of 241 biomedical image analysis challenges comprising 458 tasks across 19 imaging modalities. Our findings show substantial biases in dataset composition, including geographic location, modality-, and problem type-related biases, indicating that current benchmarks do not adequately reflect real-world clinical diversity. Despite their widespread influence, challenge datasets were frequently constrained by restrictive or ambiguous access conditions, inconsistent or non-compliant licensing practices, and incomplete documentation, limiting reproducibility and long-term reuse. Together, these shortcomings expose foundational fairness limitations in our benchmarking ecosystem and highlight a disconnect between leaderboard success and clinical relevance.
False Promises in Medical Imaging AI? Assessing Validity of Outperformance Claims
May 07, 2025Abstract:Performance comparisons are fundamental in medical imaging Artificial Intelligence (AI) research, often driving claims of superiority based on relative improvements in common performance metrics. However, such claims frequently rely solely on empirical mean performance. In this paper, we investigate whether newly proposed methods genuinely outperform the state of the art by analyzing a representative cohort of medical imaging papers. We quantify the probability of false claims based on a Bayesian approach that leverages reported results alongside empirically estimated model congruence to estimate whether the relative ranking of methods is likely to have occurred by chance. According to our results, the majority (>80%) of papers claims outperformance when introducing a new method. Our analysis further revealed a high probability (>5%) of false outperformance claims in 86% of classification papers and 53% of segmentation papers. These findings highlight a critical flaw in current benchmarking practices: claims of outperformance in medical imaging AI are frequently unsubstantiated, posing a risk of misdirecting future research efforts.
Robustness and sex differences in skin cancer detection: logistic regression vs CNNs
Apr 15, 2025
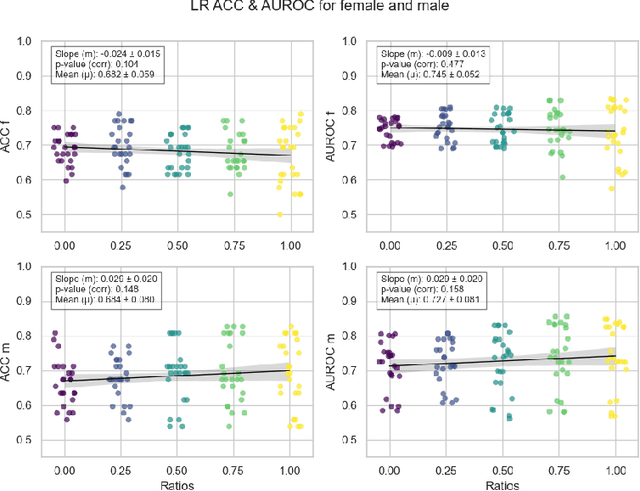
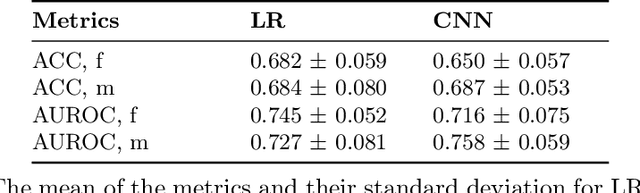
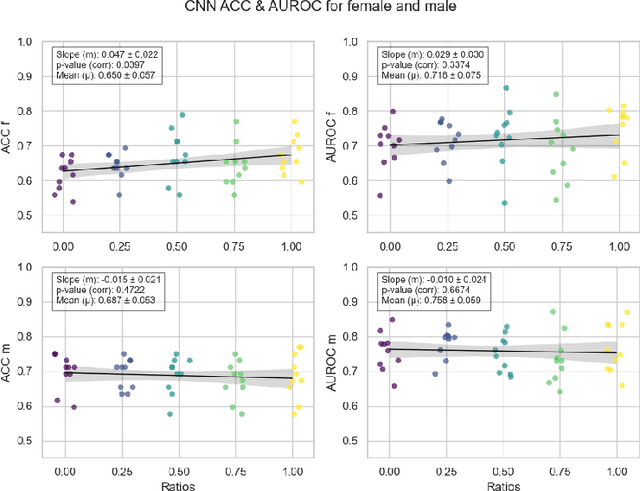
Abstract:Deep learning has been reported to achieve high performances in the detection of skin cancer, yet many challenges regarding the reproducibility of results and biases remain. This study is a replication (different data, same analysis) of a study on Alzheimer's disease [28] which studied robustness of logistic regression (LR) and convolutional neural networks (CNN) across patient sexes. We explore sex bias in skin cancer detection, using the PAD-UFES-20 dataset with LR trained on handcrafted features reflecting dermatological guidelines (ABCDE and the 7-point checklist), and a pre-trained ResNet-50 model. We evaluate these models in alignment with [28]: across multiple training datasets with varied sex composition to determine their robustness. Our results show that both the LR and the CNN were robust to the sex distributions, but the results also revealed that the CNN had a significantly higher accuracy (ACC) and area under the receiver operating characteristics (AUROC) for male patients than for female patients. We hope these findings to contribute to the growing field of investigating potential bias in popular medical machine learning methods. The data and relevant scripts to reproduce our results can be found in our Github.
Learning to Harmonize Cross-vendor X-ray Images by Non-linear Image Dynamics Correction
Apr 14, 2025Abstract:In this paper, we explore how conventional image enhancement can improve model robustness in medical image analysis. By applying commonly used normalization methods to images from various vendors and studying their influence on model generalization in transfer learning, we show that the nonlinear characteristics of domain-specific image dynamics cannot be addressed by simple linear transforms. To tackle this issue, we reformulate the image harmonization task as an exposure correction problem and propose a method termed Global Deep Curve Estimation (GDCE) to reduce domain-specific exposure mismatch. GDCE performs enhancement via a pre-defined polynomial function and is trained with the help of a ``domain discriminator'', aiming to improve model transparency in downstream tasks compared to existing black-box methods.
In the Picture: Medical Imaging Datasets, Artifacts, and their Living Review
Jan 18, 2025


Abstract:Datasets play a critical role in medical imaging research, yet issues such as label quality, shortcuts, and metadata are often overlooked. This lack of attention may harm the generalizability of algorithms and, consequently, negatively impact patient outcomes. While existing medical imaging literature reviews mostly focus on machine learning (ML) methods, with only a few focusing on datasets for specific applications, these reviews remain static -- they are published once and not updated thereafter. This fails to account for emerging evidence, such as biases, shortcuts, and additional annotations that other researchers may contribute after the dataset is published. We refer to these newly discovered findings of datasets as research artifacts. To address this gap, we propose a living review that continuously tracks public datasets and their associated research artifacts across multiple medical imaging applications. Our approach includes a framework for the living review to monitor data documentation artifacts, and an SQL database to visualize the citation relationships between research artifact and dataset. Lastly, we discuss key considerations for creating medical imaging datasets, review best practices for data annotation, discuss the significance of shortcuts and demographic diversity, and emphasize the importance of managing datasets throughout their entire lifecycle. Our demo is publicly available at http://130.226.140.142.
On dataset transferability in medical image classification
Dec 28, 2024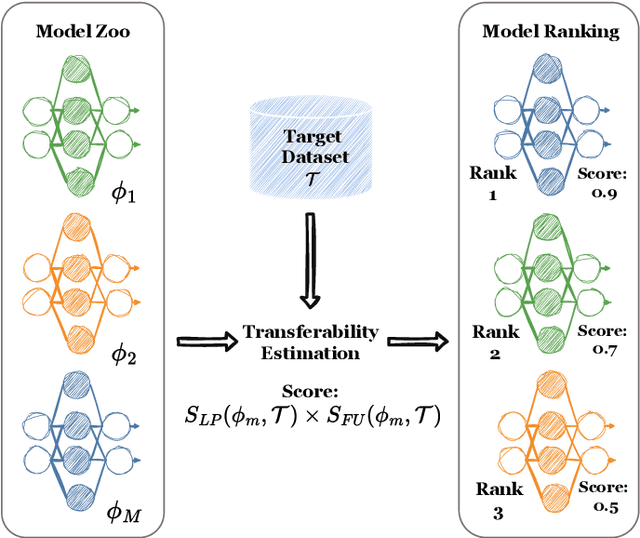
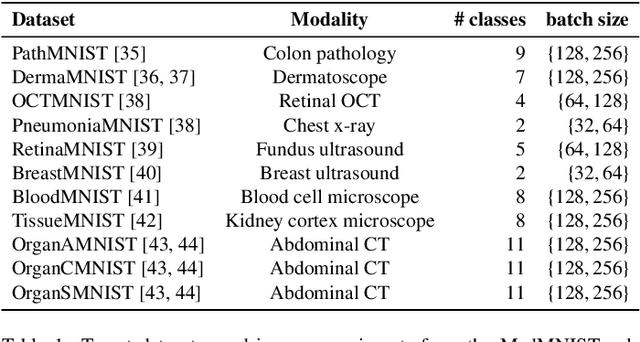
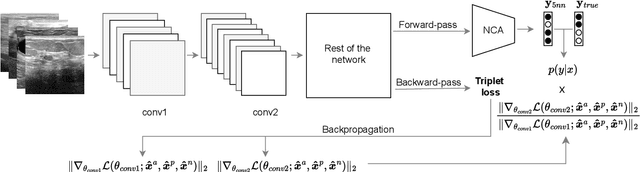
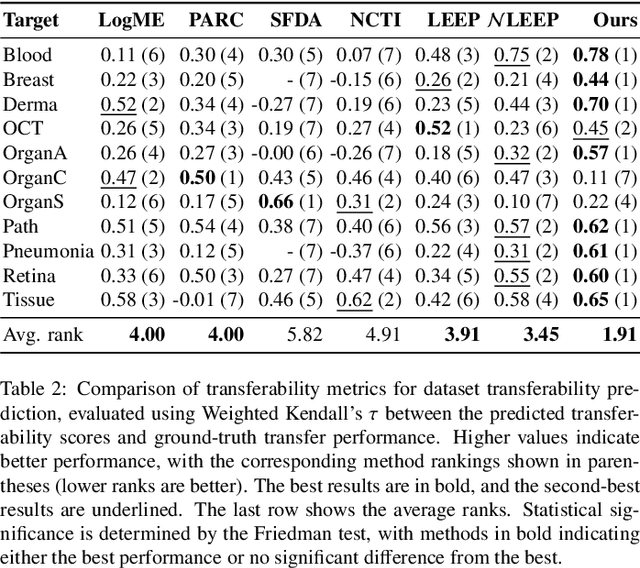
Abstract:Current transferability estimation methods designed for natural image datasets are often suboptimal in medical image classification. These methods primarily focus on estimating the suitability of pre-trained source model features for a target dataset, which can lead to unrealistic predictions, such as suggesting that the target dataset is the best source for itself. To address this, we propose a novel transferability metric that combines feature quality with gradients to evaluate both the suitability and adaptability of source model features for target tasks. We evaluate our approach in two new scenarios: source dataset transferability for medical image classification and cross-domain transferability. Our results show that our method outperforms existing transferability metrics in both settings. We also provide insight into the factors influencing transfer performance in medical image classification, as well as the dynamics of cross-domain transfer from natural to medical images. Additionally, we provide ground-truth transfer performance benchmarking results to encourage further research into transferability estimation for medical image classification. Our code and experiments are available at https://github.com/DovileDo/transferability-in-medical-imaging.
SeagrassFinder: Deep Learning for Eelgrass Detection and Coverage Estimation in the Wild
Dec 20, 2024



Abstract:Seagrass meadows play a crucial role in marine ecosystems, providing important services such as carbon sequestration, water quality improvement, and habitat provision. Monitoring the distribution and abundance of seagrass is essential for environmental impact assessments and conservation efforts. However, the current manual methods of analyzing underwater video transects to assess seagrass coverage are time-consuming and subjective. This work explores the use of deep learning models to automate the process of seagrass detection and coverage estimation from underwater video data. A dataset of over 8,300 annotated underwater images was created, and several deep learning architectures, including ResNet, InceptionNetV3, DenseNet, and Vision Transformer, were evaluated for the task of binary classification of ``Eelgrass Present'' and ``Eelgrass Absent'' images. The results demonstrate that deep learning models, particularly the Vision Transformer, can achieve high performance in predicting eelgrass presence, with AUROC scores exceeding 0.95 on the final test dataset. The use of transfer learning and the application of the Deep WaveNet underwater image enhancement model further improved the models' capabilities. The proposed methodology allows for the efficient processing of large volumes of video data, enabling the acquisition of much more detailed information on seagrass distributions compared to current manual methods. This information is crucial for environmental impact assessments and monitoring programs, as seagrasses are important indicators of coastal ecosystem health. Overall, this project demonstrates the value that deep learning can bring to the field of marine ecology and environmental monitoring.
Mask of truth: model sensitivity to unexpected regions of medical images
Dec 05, 2024Abstract:The development of larger models for medical image analysis has led to increased performance. However, it also affected our ability to explain and validate model decisions. Models can use non-relevant parts of images, also called spurious correlations or shortcuts, to obtain high performance on benchmark datasets but fail in real-world scenarios. In this work, we challenge the capacity of convolutional neural networks (CNN) to classify chest X-rays and eye fundus images while masking out clinically relevant parts of the image. We show that all models trained on the PadChest dataset, irrespective of the masking strategy, are able to obtain an Area Under the Curve (AUC) above random. Moreover, the models trained on full images obtain good performance on images without the region of interest (ROI), even superior to the one obtained on images only containing the ROI. We also reveal a possible spurious correlation in the Chaksu dataset while the performances are more aligned with the expectation of an unbiased model. We go beyond the performance analysis with the usage of the explainability method SHAP and the analysis of embeddings. We asked a radiology resident to interpret chest X-rays under different masking to complement our findings with clinical knowledge. Our code is available at https://github.com/TheoSourget/MMC_Masking and https://github.com/TheoSourget/MMC_Masking_EyeFundus
Confidence intervals uncovered: Are we ready for real-world medical imaging AI?
Sep 27, 2024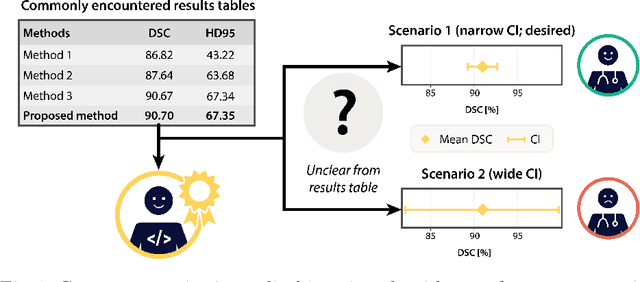
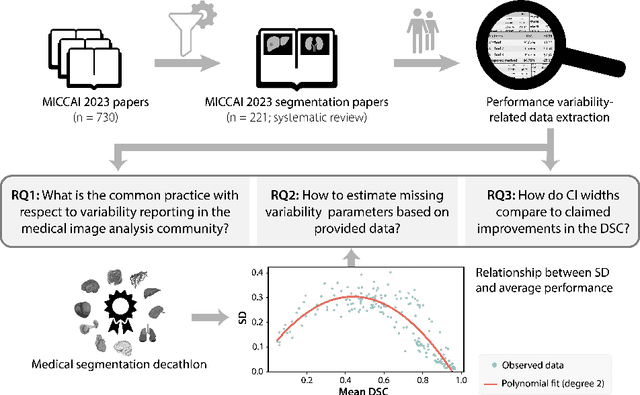
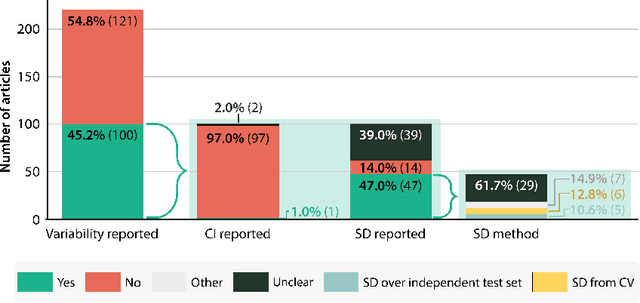

Abstract:Medical imaging is spearheading the AI transformation of healthcare. Performance reporting is key to determine which methods should be translated into clinical practice. Frequently, broad conclusions are simply derived from mean performance values. In this paper, we argue that this common practice is often a misleading simplification as it ignores performance variability. Our contribution is threefold. (1) Analyzing all MICCAI segmentation papers (n = 221) published in 2023, we first observe that more than 50% of papers do not assess performance variability at all. Moreover, only one (0.5%) paper reported confidence intervals (CIs) for model performance. (2) To address the reporting bottleneck, we show that the unreported standard deviation (SD) in segmentation papers can be approximated by a second-order polynomial function of the mean Dice similarity coefficient (DSC). Based on external validation data from 56 previous MICCAI challenges, we demonstrate that this approximation can accurately reconstruct the CI of a method using information provided in publications. (3) Finally, we reconstructed 95% CIs around the mean DSC of MICCAI 2023 segmentation papers. The median CI width was 0.03 which is three times larger than the median performance gap between the first and second ranked method. For more than 60% of papers, the mean performance of the second-ranked method was within the CI of the first-ranked method. We conclude that current publications typically do not provide sufficient evidence to support which models could potentially be translated into clinical practice.
Dataset Distribution Impacts Model Fairness: Single vs. Multi-Task Learning
Jul 24, 2024Abstract:The influence of bias in datasets on the fairness of model predictions is a topic of ongoing research in various fields. We evaluate the performance of skin lesion classification using ResNet-based CNNs, focusing on patient sex variations in training data and three different learning strategies. We present a linear programming method for generating datasets with varying patient sex and class labels, taking into account the correlations between these variables. We evaluated the model performance using three different learning strategies: a single-task model, a reinforcing multi-task model, and an adversarial learning scheme. Our observations include: 1) sex-specific training data yields better results, 2) single-task models exhibit sex bias, 3) the reinforcement approach does not remove sex bias, 4) the adversarial model eliminates sex bias in cases involving only female patients, and 5) datasets that include male patients enhance model performance for the male subgroup, even when female patients are the majority. To generalise these findings, in future research, we will examine more demographic attributes, like age, and other possibly confounding factors, such as skin colour and artefacts in the skin lesions. We make all data and models available on GitHub.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge