Aasa Feragen
Weight Space Correlation Analysis: Quantifying Feature Utilization in Deep Learning Models
Dec 15, 2025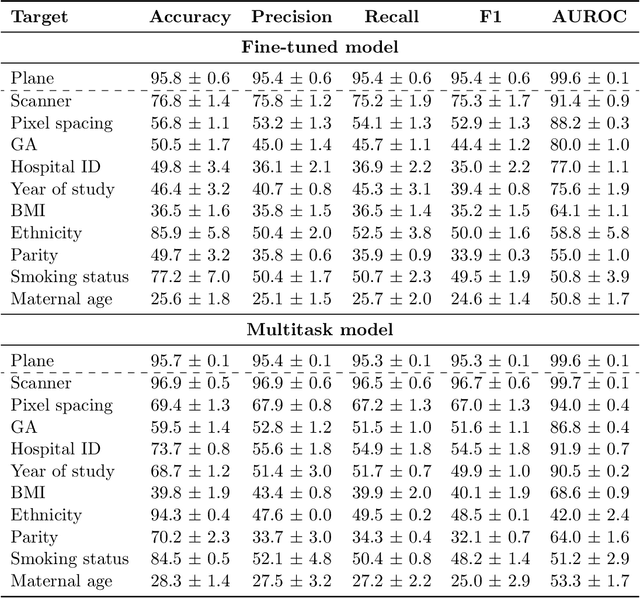
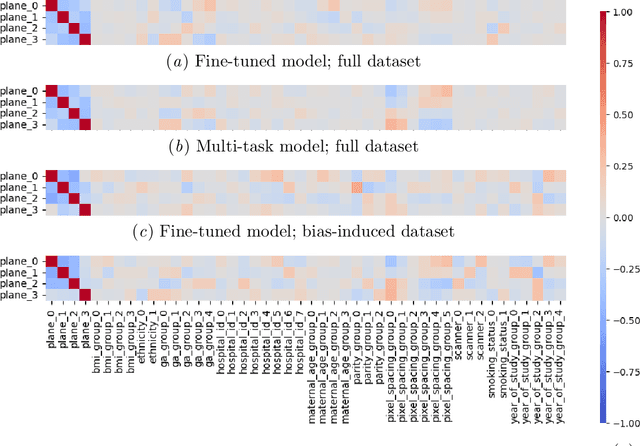
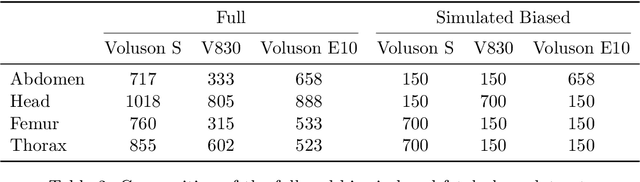

Abstract:Deep learning models in medical imaging are susceptible to shortcut learning, relying on confounding metadata (e.g., scanner model) that is often encoded in image embeddings. The crucial question is whether the model actively utilizes this encoded information for its final prediction. We introduce Weight Space Correlation Analysis, an interpretable methodology that quantifies feature utilization by measuring the alignment between the classification heads of a primary clinical task and auxiliary metadata tasks. We first validate our method by successfully detecting artificially induced shortcut learning. We then apply it to probe the feature utilization of an SA-SonoNet model trained for Spontaneous Preterm Birth (sPTB) prediction. Our analysis confirmed that while the embeddings contain substantial metadata, the sPTB classifier's weight vectors were highly correlated with clinically relevant factors (e.g., birth weight) but decoupled from clinically irrelevant acquisition factors (e.g. scanner). Our methodology provides a tool to verify model trustworthiness, demonstrating that, in the absence of induced bias, the clinical model selectively utilizes features related to the genuine clinical signal.
Who Does Your Algorithm Fail? Investigating Age and Ethnic Bias in the MAMA-MIA Dataset
Oct 31, 2025Abstract:Deep learning models aim to improve diagnostic workflows, but fairness evaluation remains underexplored beyond classification, e.g., in image segmentation. Unaddressed segmentation bias can lead to disparities in the quality of care for certain populations, potentially compounded across clinical decision points and amplified through iterative model development. Here, we audit the fairness of the automated segmentation labels provided in the breast cancer tumor segmentation dataset MAMA-MIA. We evaluate automated segmentation quality across age, ethnicity, and data source. Our analysis reveals an intrinsic age-related bias against younger patients that continues to persist even after controlling for confounding factors, such as data source. We hypothesize that this bias may be linked to physiological factors, a known challenge for both radiologists and automated systems. Finally, we show how aggregating data from multiple data sources influences site-specific ethnic biases, underscoring the necessity of investigating data at a granular level.
General Methods Make Great Domain-specific Foundation Models: A Case-study on Fetal Ultrasound
Jun 24, 2025Abstract:With access to large-scale, unlabeled medical datasets, researchers are confronted with two questions: Should they attempt to pretrain a custom foundation model on this medical data, or use transfer-learning from an existing generalist model? And, if a custom model is pretrained, are novel methods required? In this paper we explore these questions by conducting a case-study, in which we train a foundation model on a large regional fetal ultrasound dataset of 2M images. By selecting the well-established DINOv2 method for pretraining, we achieve state-of-the-art results on three fetal ultrasound datasets, covering data from different countries, classification, segmentation, and few-shot tasks. We compare against a series of models pretrained on natural images, ultrasound images, and supervised baselines. Our results demonstrate two key insights: (i) Pretraining on custom data is worth it, even if smaller models are trained on less data, as scaling in natural image pretraining does not translate to ultrasound performance. (ii) Well-tuned methods from computer vision are making it feasible to train custom foundation models for a given medical domain, requiring no hyperparameter tuning and little methodological adaptation. Given these findings, we argue that a bias towards methodological innovation should be avoided when developing domain specific foundation models under common computational resource constraints.
Patronus: Bringing Transparency to Diffusion Models with Prototypes
Mar 28, 2025Abstract:Diffusion-based generative models, such as Denoising Diffusion Probabilistic Models (DDPMs), have achieved remarkable success in image generation, but their step-by-step denoising process remains opaque, leaving critical aspects of the generation mechanism unexplained. To address this, we introduce \emph{Patronus}, an interpretable diffusion model inspired by ProtoPNet. Patronus integrates a prototypical network into DDPMs, enabling the extraction of prototypes and conditioning of the generation process on their prototype activation vector. This design enhances interpretability by showing the learned prototypes and how they influence the generation process. Additionally, the model supports downstream tasks like image manipulation, enabling more transparent and controlled modifications. Moreover, Patronus could reveal shortcut learning in the generation process by detecting unwanted correlations between learned prototypes. Notably, Patronus operates entirely without any annotations or text prompts. This work opens new avenues for understanding and controlling diffusion models through prototype-based interpretability. Our code is available at \href{https://github.com/nina-weng/patronus}{https://github.com/nina-weng/patronus}.
Are generative models fair? A study of racial bias in dermatological image generation
Jan 20, 2025



Abstract:Racial bias in medicine, particularly in dermatology, presents significant ethical and clinical challenges. It often results from the underrepresentation of darker skin tones in training datasets for machine learning models. While efforts to address bias in dermatology have focused on improving dataset diversity and mitigating disparities in discriminative models, the impact of racial bias on generative models remains underexplored. Generative models, such as Variational Autoencoders (VAEs), are increasingly used in healthcare applications, yet their fairness across diverse skin tones is currently not well understood. In this study, we evaluate the fairness of generative models in clinical dermatology with respect to racial bias. For this purpose, we first train a VAE with a perceptual loss to generate and reconstruct high-quality skin images across different skin tones. We utilize the Fitzpatrick17k dataset to examine how racial bias influences the representation and performance of these models. Our findings indicate that the VAE is influenced by the diversity of skin tones in the training dataset, with better performance observed for lighter skin tones. Additionally, the uncertainty estimates produced by the VAE are ineffective in assessing the model's fairness. These results highlight the need for improved uncertainty quantification mechanisms to detect and address racial bias in generative models for trustworthy healthcare technologies.
In the Picture: Medical Imaging Datasets, Artifacts, and their Living Review
Jan 18, 2025


Abstract:Datasets play a critical role in medical imaging research, yet issues such as label quality, shortcuts, and metadata are often overlooked. This lack of attention may harm the generalizability of algorithms and, consequently, negatively impact patient outcomes. While existing medical imaging literature reviews mostly focus on machine learning (ML) methods, with only a few focusing on datasets for specific applications, these reviews remain static -- they are published once and not updated thereafter. This fails to account for emerging evidence, such as biases, shortcuts, and additional annotations that other researchers may contribute after the dataset is published. We refer to these newly discovered findings of datasets as research artifacts. To address this gap, we propose a living review that continuously tracks public datasets and their associated research artifacts across multiple medical imaging applications. Our approach includes a framework for the living review to monitor data documentation artifacts, and an SQL database to visualize the citation relationships between research artifact and dataset. Lastly, we discuss key considerations for creating medical imaging datasets, review best practices for data annotation, discuss the significance of shortcuts and demographic diversity, and emphasize the importance of managing datasets throughout their entire lifecycle. Our demo is publicly available at http://130.226.140.142.
Graph Counterfactual Explainable AI via Latent Space Traversal
Jan 15, 2025Abstract:Explaining the predictions of a deep neural network is a nontrivial task, yet high-quality explanations for predictions are often a prerequisite for practitioners to trust these models. Counterfactual explanations aim to explain predictions by finding the ''nearest'' in-distribution alternative input whose prediction changes in a pre-specified way. However, it remains an open question how to define this nearest alternative input, whose solution depends on both the domain (e.g. images, graphs, tabular data, etc.) and the specific application considered. For graphs, this problem is complicated i) by their discrete nature, as opposed to the continuous nature of state-of-the-art graph classifiers; and ii) by the node permutation group acting on the graphs. We propose a method to generate counterfactual explanations for any differentiable black-box graph classifier, utilizing a case-specific permutation equivariant graph variational autoencoder. We generate counterfactual explanations in a continuous fashion by traversing the latent space of the autoencoder across the classification boundary of the classifier, allowing for seamless integration of discrete graph structure and continuous graph attributes. We empirically validate the approach on three graph datasets, showing that our model is consistently high-performing and more robust than the baselines.
* Published at Northern Lights Deep Learning Conference 2025
Counterfactual Explanations via Riemannian Latent Space Traversal
Nov 04, 2024Abstract:The adoption of increasingly complex deep models has fueled an urgent need for insight into how these models make predictions. Counterfactual explanations form a powerful tool for providing actionable explanations to practitioners. Previously, counterfactual explanation methods have been designed by traversing the latent space of generative models. Yet, these latent spaces are usually greatly simplified, with most of the data distribution complexity contained in the decoder rather than the latent embedding. Thus, traversing the latent space naively without taking the nonlinear decoder into account can lead to unnatural counterfactual trajectories. We introduce counterfactual explanations obtained using a Riemannian metric pulled back via the decoder and the classifier under scrutiny. This metric encodes information about the complex geometric structure of the data and the learned representation, enabling us to obtain robust counterfactual trajectories with high fidelity, as demonstrated by our experiments in real-world tabular datasets.
Unsupervised Detection of Fetal Brain Anomalies using Denoising Diffusion Models
Aug 07, 2024Abstract:Congenital malformations of the brain are among the most common fetal abnormalities that impact fetal development. Previous anomaly detection methods on ultrasound images are based on supervised learning, rely on manual annotations, and risk missing underrepresented categories. In this work, we frame fetal brain anomaly detection as an unsupervised task using diffusion models. To this end, we employ an inpainting-based Noise Agnostic Anomaly Detection approach that identifies the abnormality using diffusion-reconstructed fetal brain images from multiple noise levels. Our approach only requires normal fetal brain ultrasound images for training, addressing the limited availability of abnormal data. Our experiments on a real-world clinical dataset show the potential of using unsupervised methods for fetal brain anomaly detection. Additionally, we comprehensively evaluate how different noise types affect diffusion models in the fetal anomaly detection domain.
Navigating Uncertainty in Medical Image Segmentation
Jul 23, 2024



Abstract:We address the selection and evaluation of uncertain segmentation methods in medical imaging and present two case studies: prostate segmentation, illustrating that for minimal annotator variation simple deterministic models can suffice, and lung lesion segmentation, highlighting the limitations of the Generalized Energy Distance (GED) in model selection. Our findings lead to guidelines for accurately choosing and developing uncertain segmentation models, that integrate aleatoric and epistemic components. These guidelines are designed to aid researchers and practitioners in better developing, selecting, and evaluating uncertain segmentation methods, thereby facilitating enhanced adoption and effective application of segmentation uncertainty in practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge