Judy Wawira Gichoya
Subgroup Performance of a Commercial Digital Breast Tomosynthesis Model for Breast Cancer Detection
Mar 17, 2025Abstract:While research has established the potential of AI models for mammography to improve breast cancer screening outcomes, there have not been any detailed subgroup evaluations performed to assess the strengths and weaknesses of commercial models for digital breast tomosynthesis (DBT) imaging. This study presents a granular evaluation of the Lunit INSIGHT DBT model on a large retrospective cohort of 163,449 screening mammography exams from the Emory Breast Imaging Dataset (EMBED). Model performance was evaluated in a binary context with various negative exam types (162,081 exams) compared against screen detected cancers (1,368 exams) as the positive class. The analysis was stratified across demographic, imaging, and pathologic subgroups to identify potential disparities. The model achieved an overall AUC of 0.91 (95% CI: 0.90-0.92) with a precision of 0.08 (95% CI: 0.08-0.08), and a recall of 0.73 (95% CI: 0.71-0.76). Performance was found to be robust across demographics, but cases with non-invasive cancers (AUC: 0.85, 95% CI: 0.83-0.87), calcifications (AUC: 0.80, 95% CI: 0.78-0.82), and dense breast tissue (AUC: 0.90, 95% CI: 0.88-0.91) were associated with significantly lower performance compared to other groups. These results highlight the need for detailed evaluation of model characteristics and vigilance in considering adoption of new tools for clinical deployment.
In the Picture: Medical Imaging Datasets, Artifacts, and their Living Review
Jan 18, 2025


Abstract:Datasets play a critical role in medical imaging research, yet issues such as label quality, shortcuts, and metadata are often overlooked. This lack of attention may harm the generalizability of algorithms and, consequently, negatively impact patient outcomes. While existing medical imaging literature reviews mostly focus on machine learning (ML) methods, with only a few focusing on datasets for specific applications, these reviews remain static -- they are published once and not updated thereafter. This fails to account for emerging evidence, such as biases, shortcuts, and additional annotations that other researchers may contribute after the dataset is published. We refer to these newly discovered findings of datasets as research artifacts. To address this gap, we propose a living review that continuously tracks public datasets and their associated research artifacts across multiple medical imaging applications. Our approach includes a framework for the living review to monitor data documentation artifacts, and an SQL database to visualize the citation relationships between research artifact and dataset. Lastly, we discuss key considerations for creating medical imaging datasets, review best practices for data annotation, discuss the significance of shortcuts and demographic diversity, and emphasize the importance of managing datasets throughout their entire lifecycle. Our demo is publicly available at http://130.226.140.142.
Benchmarking bias: Expanding clinical AI model card to incorporate bias reporting of social and non-social factors
Nov 21, 2023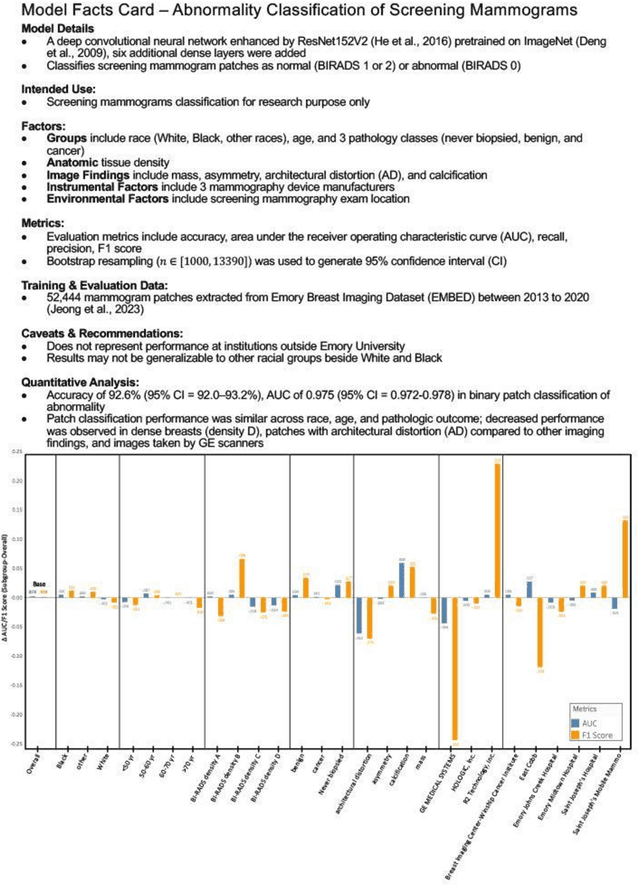
Abstract:Clinical AI model reporting cards should be expanded to incorporate a broad bias reporting of both social and non-social factors. Non-social factors consider the role of other factors, such as disease dependent, anatomic, or instrument factors on AI model bias, which are essential to ensure safe deployment.
Performance Gaps of Artificial Intelligence Models Screening Mammography -- Towards Fair and Interpretable Models
May 08, 2023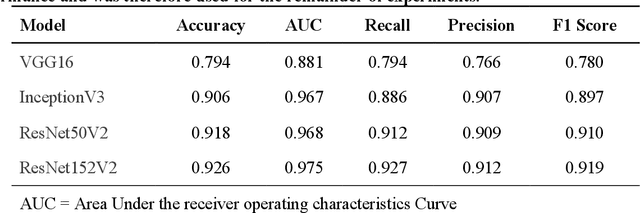
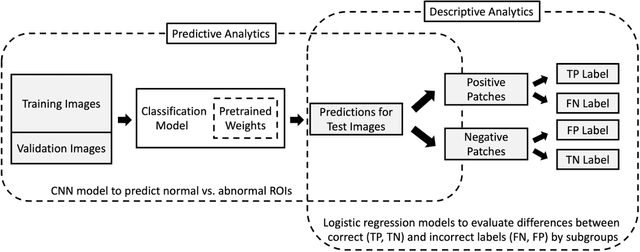
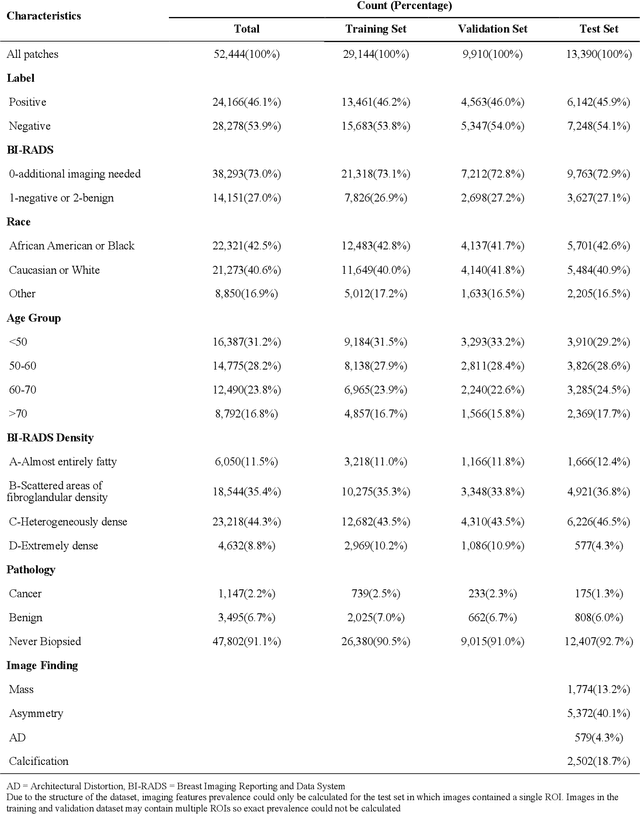
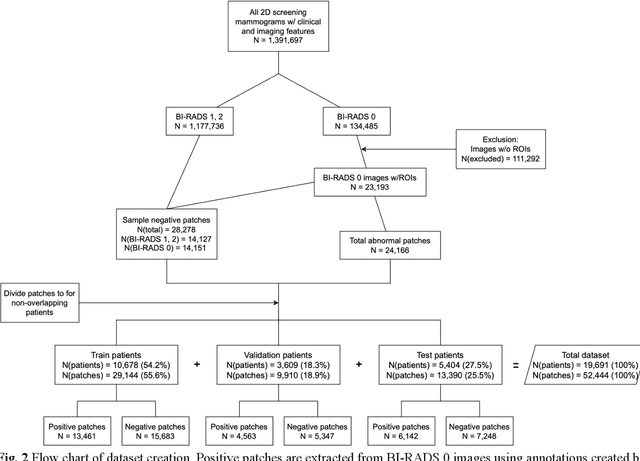
Abstract:Purpose: To analyze the demographic and imaging characteristics associated with increased risk of failure for abnormality classification in screening mammograms. Materials and Methods: This retrospective study used data from the Emory BrEast Imaging Dataset (EMBED) which includes mammograms from 115,931 patients imaged at Emory University Healthcare between 2013 to 2020. Clinical and imaging data includes Breast Imaging Reporting and Data System (BI-RADS) assessment, region of interest coordinates for abnormalities, imaging features, pathologic outcomes, and patient demographics. Multiple deep learning models were developed to distinguish between patches of abnormal tissue and randomly selected patches of normal tissue from the screening mammograms. We assessed model performance overall and within subgroups defined by age, race, pathologic outcome, and imaging characteristics to evaluate reasons for misclassifications. Results: On a test set size of 5,810 studies (13,390 patches), a ResNet152V2 model trained to classify normal versus abnormal tissue patches achieved an accuracy of 92.6% (95% CI = 92.0-93.2%), and area under the receiver operative characteristics curve 0.975 (95% CI = 0.972-0.978). Imaging characteristics associated with higher misclassifications of images include higher tissue densities (risk ratio [RR]=1.649; p=.010, BI-RADS density C and RR=2.026; p=.003, BI-RADS density D), and presence of architectural distortion (RR=1.026; p<.001). Conclusion: Even though deep learning models for abnormality classification can perform well in screening mammography, we demonstrate certain imaging features that result in worse model performance. This is the first such work to systematically evaluate breast abnormality classification by various subgroups and better-informed developers and end-users of population subgroups which are likely to experience biased model performance.
Report of the Medical Image De-Identification Task Group -- Best Practices and Recommendations
Apr 01, 2023Abstract:This report addresses the technical aspects of de-identification of medical images of human subjects and biospecimens, such that re-identification risk of ethical, moral, and legal concern is sufficiently reduced to allow unrestricted public sharing for any purpose, regardless of the jurisdiction of the source and distribution sites. All medical images, regardless of the mode of acquisition, are considered, though the primary emphasis is on those with accompanying data elements, especially those encoded in formats in which the data elements are embedded, particularly Digital Imaging and Communications in Medicine (DICOM). These images include image-like objects such as Segmentations, Parametric Maps, and Radiotherapy (RT) Dose objects. The scope also includes related non-image objects, such as RT Structure Sets, Plans and Dose Volume Histograms, Structured Reports, and Presentation States. Only de-identification of publicly released data is considered, and alternative approaches to privacy preservation, such as federated learning for artificial intelligence (AI) model development, are out of scope, as are issues of privacy leakage from AI model sharing. Only technical issues of public sharing are addressed.
Augmenting Vision Language Pretraining by Learning Codebook with Visual Semantics
Jul 31, 2022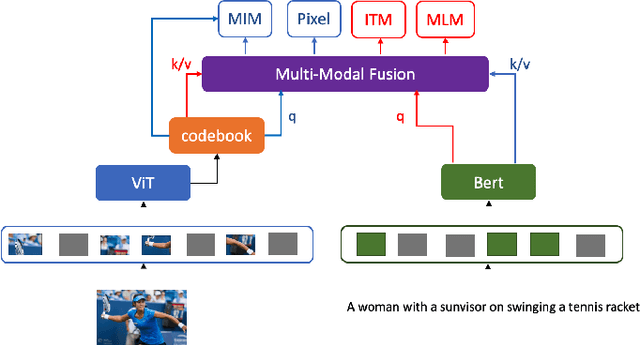
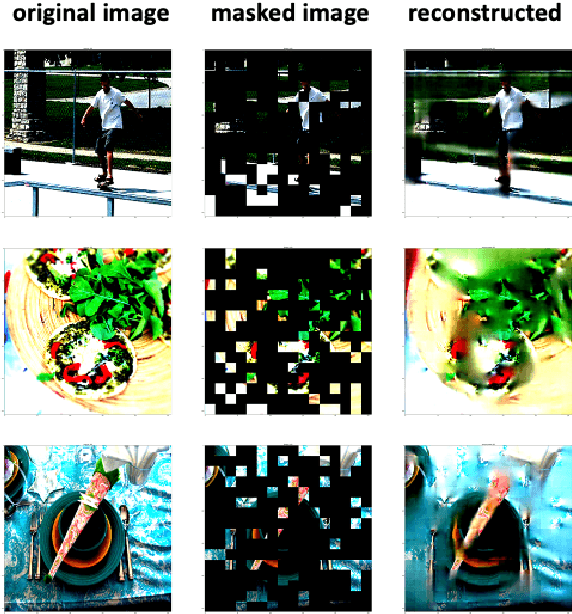


Abstract:Language modality within the vision language pretraining framework is innately discretized, endowing each word in the language vocabulary a semantic meaning. In contrast, visual modality is inherently continuous and high-dimensional, which potentially prohibits the alignment as well as fusion between vision and language modalities. We therefore propose to "discretize" the visual representation by joint learning a codebook that imbues each visual token a semantic. We then utilize these discretized visual semantics as self-supervised ground-truths for building our Masked Image Modeling objective, a counterpart of Masked Language Modeling which proves successful for language models. To optimize the codebook, we extend the formulation of VQ-VAE which gives a theoretic guarantee. Experiments validate the effectiveness of our approach across common vision-language benchmarks.
Few-Shot Transfer Learning to improve Chest X-Ray pathology detection using limited triplets
Apr 16, 2022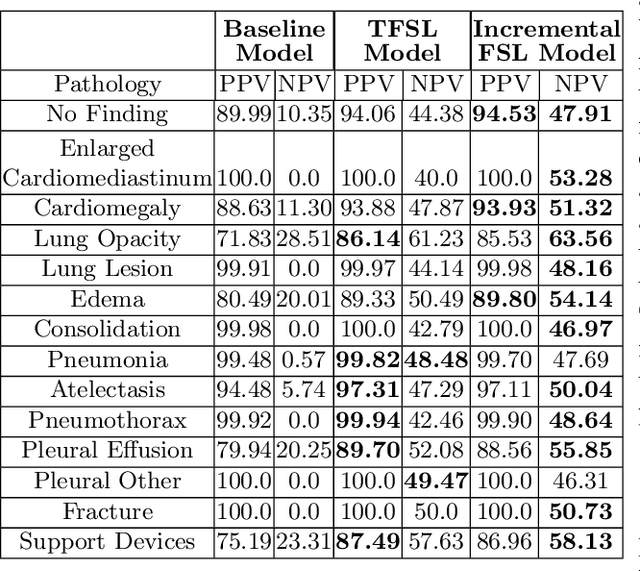
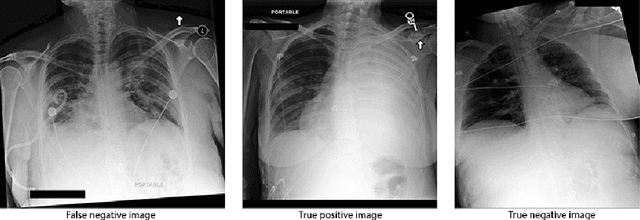
Abstract:Deep learning approaches applied to medical imaging have reached near-human or better-than-human performance on many diagnostic tasks. For instance, the CheXpert competition on detecting pathologies in chest x-rays has shown excellent multi-class classification performance. However, training and validating deep learning models require extensive collections of images and still produce false inferences, as identified by a human-in-the-loop. In this paper, we introduce a practical approach to improve the predictions of a pre-trained model through Few-Shot Learning (FSL). After training and validating a model, a small number of false inference images are collected to retrain the model using \textbf{\textit{Image Triplets}} - a false positive or false negative, a true positive, and a true negative. The retrained FSL model produces considerable gains in performance with only a few epochs and few images. In addition, FSL opens rapid retraining opportunities for human-in-the-loop systems, where a radiologist can relabel false inferences, and the model can be quickly retrained. We compare our retrained model performance with existing FSL approaches in medical imaging that train and evaluate models at once.
OSCARS: An Outlier-Sensitive Content-Based Radiography Retrieval System
Apr 06, 2022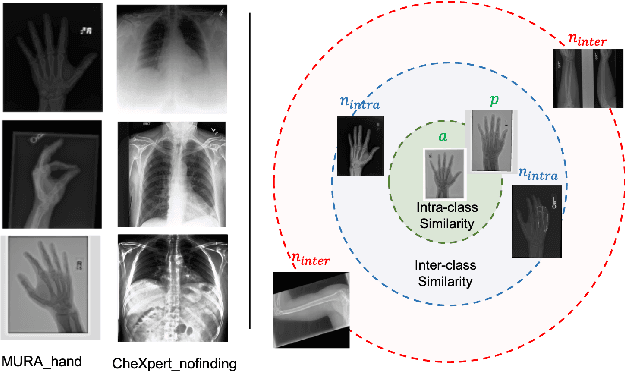

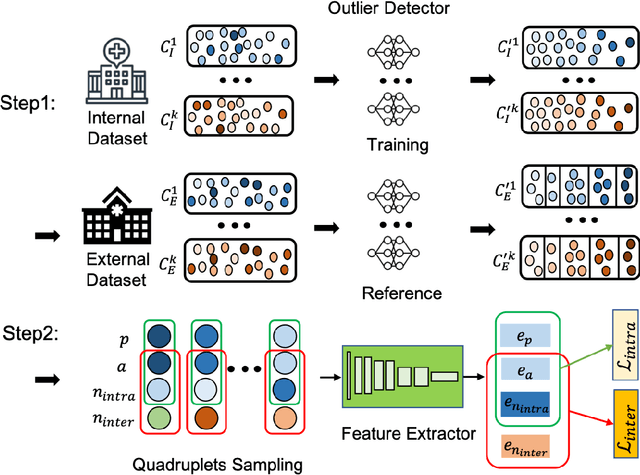
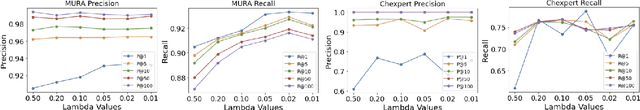
Abstract:Improving the retrieval relevance on noisy datasets is an emerging need for the curation of a large-scale clean dataset in the medical domain. While existing methods can be applied for class-wise retrieval (aka. inter-class), they cannot distinguish the granularity of likeness within the same class (aka. intra-class). The problem is exacerbated on medical external datasets, where noisy samples of the same class are treated equally during training. Our goal is to identify both intra/inter-class similarities for fine-grained retrieval. To achieve this, we propose an Outlier-Sensitive Content-based rAdiologhy Retrieval System (OSCARS), consisting of two steps. First, we train an outlier detector on a clean internal dataset in an unsupervised manner. Then we use the trained detector to generate the anomaly scores on the external dataset, whose distribution will be used to bin intra-class variations. Second, we propose a quadruplet (a, p, nintra, ninter) sampling strategy, where intra-class negatives nintra are sampled from bins of the same class other than the bin anchor a belongs to, while niner are randomly sampled from inter-classes. We suggest a weighted metric learning objective to balance the intra and inter-class feature learning. We experimented on two representative public radiography datasets. Experiments show the effectiveness of our approach. The training and evaluation code can be found in https://github.com/XiaoyuanGuo/oscars.
Best Practices and Scoring System on Reviewing A.I. based Medical Imaging Papers: Part 1 Classification
Feb 03, 2022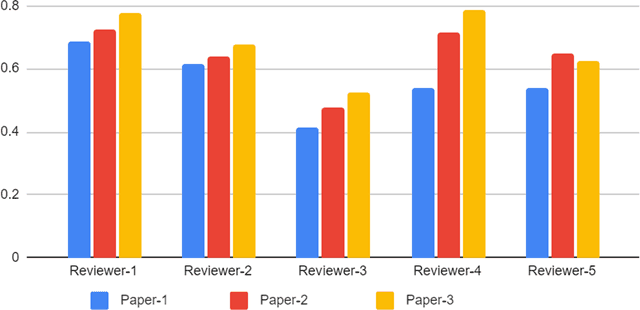
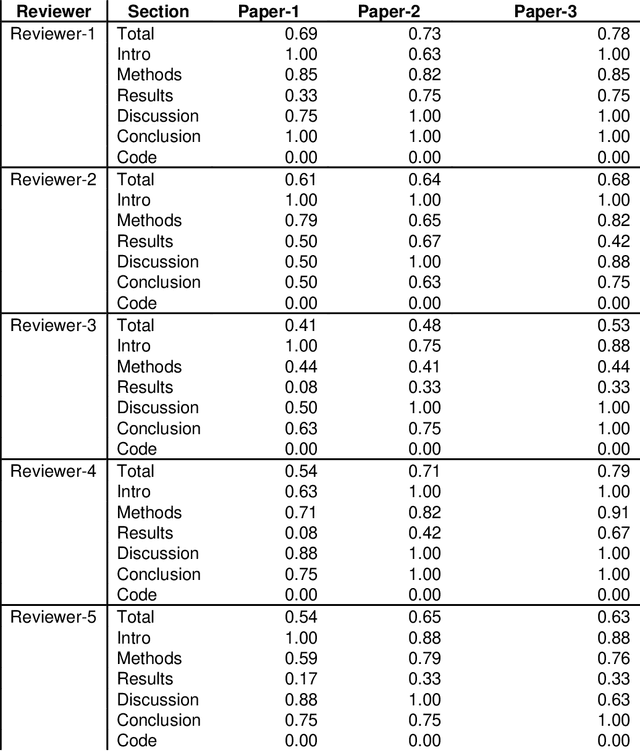
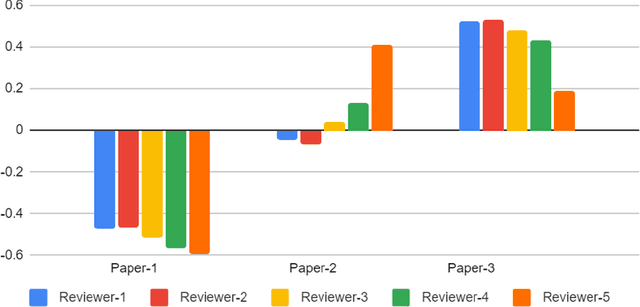
Abstract:With the recent advances in A.I. methodologies and their application to medical imaging, there has been an explosion of related research programs utilizing these techniques to produce state-of-the-art classification performance. Ultimately, these research programs culminate in submission of their work for consideration in peer reviewed journals. To date, the criteria for acceptance vs. rejection is often subjective; however, reproducible science requires reproducible review. The Machine Learning Education Sub-Committee of SIIM has identified a knowledge gap and a serious need to establish guidelines for reviewing these studies. Although there have been several recent papers with this goal, this present work is written from the machine learning practitioners standpoint. In this series, the committee will address the best practices to be followed in an A.I.-based study and present the required sections in terms of examples and discussion of what should be included to make the studies cohesive, reproducible, accurate, and self-contained. This first entry in the series focuses on the task of image classification. Elements such as dataset curation, data pre-processing steps, defining an appropriate reference standard, data partitioning, model architecture and training are discussed. The sections are presented as they would be detailed in a typical manuscript, with content describing the necessary information that should be included to make sure the study is of sufficient quality to be considered for publication. The goal of this series is to provide resources to not only help improve the review process for A.I.-based medical imaging papers, but to facilitate a standard for the information that is presented within all components of the research study. We hope to provide quantitative metrics in what otherwise may be a qualitative review process.
MedShift: identifying shift data for medical dataset curation
Dec 27, 2021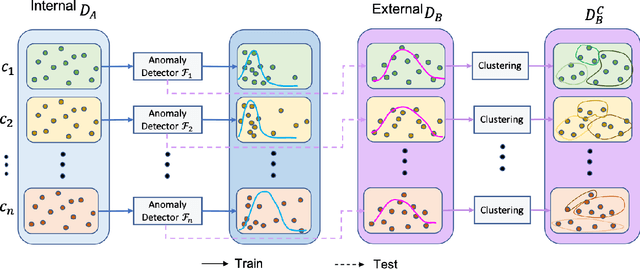
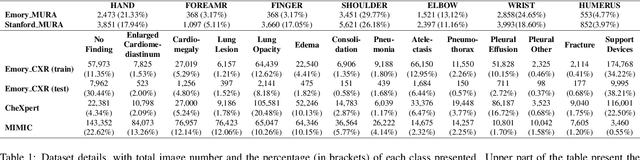
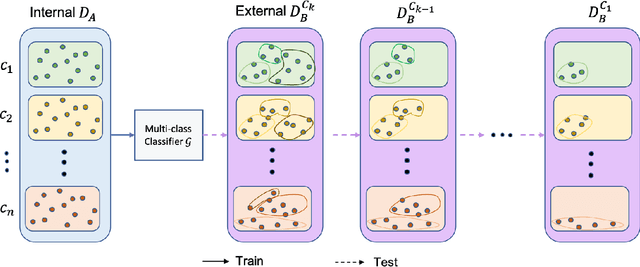
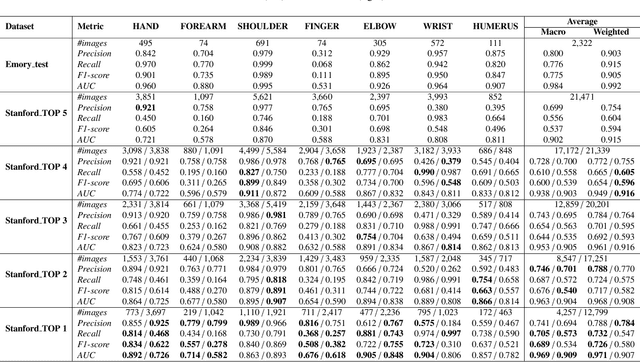
Abstract:To curate a high-quality dataset, identifying data variance between the internal and external sources is a fundamental and crucial step. However, methods to detect shift or variance in data have not been significantly researched. Challenges to this are the lack of effective approaches to learn dense representation of a dataset and difficulties of sharing private data across medical institutions. To overcome the problems, we propose a unified pipeline called MedShift to detect the top-level shift samples and thus facilitate the medical curation. Given an internal dataset A as the base source, we first train anomaly detectors for each class of dataset A to learn internal distributions in an unsupervised way. Second, without exchanging data across sources, we run the trained anomaly detectors on an external dataset B for each class. The data samples with high anomaly scores are identified as shift data. To quantify the shiftness of the external dataset, we cluster B's data into groups class-wise based on the obtained scores. We then train a multi-class classifier on A and measure the shiftness with the classifier's performance variance on B by gradually dropping the group with the largest anomaly score for each class. Additionally, we adapt a dataset quality metric to help inspect the distribution differences for multiple medical sources. We verify the efficacy of MedShift with musculoskeletal radiographs (MURA) and chest X-rays datasets from more than one external source. Experiments show our proposed shift data detection pipeline can be beneficial for medical centers to curate high-quality datasets more efficiently. An interface introduction video to visualize our results is available at https://youtu.be/V3BF0P1sxQE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge