Hari Trivedi
RSNA Large Language Model Benchmark Dataset for Chest Radiographs of Cardiothoracic Disease: Radiologist Evaluation and Validation Enhanced by AI Labels (REVEAL-CXR)
Jan 21, 2026Abstract:Multimodal large language models have demonstrated comparable performance to that of radiology trainees on multiple-choice board-style exams. However, to develop clinically useful multimodal LLM tools, high-quality benchmarks curated by domain experts are essential. To curate released and holdout datasets of 100 chest radiographic studies each and propose an artificial intelligence (AI)-assisted expert labeling procedure to allow radiologists to label studies more efficiently. A total of 13,735 deidentified chest radiographs and their corresponding reports from the MIDRC were used. GPT-4o extracted abnormal findings from the reports, which were then mapped to 12 benchmark labels with a locally hosted LLM (Phi-4-Reasoning). From these studies, 1,000 were sampled on the basis of the AI-suggested benchmark labels for expert review; the sampling algorithm ensured that the selected studies were clinically relevant and captured a range of difficulty levels. Seventeen chest radiologists participated, and they marked "Agree all", "Agree mostly" or "Disagree" to indicate their assessment of the correctness of the LLM suggested labels. Each chest radiograph was evaluated by three experts. Of these, at least two radiologists selected "Agree All" for 381 radiographs. From this set, 200 were selected, prioritizing those with less common or multiple finding labels, and divided into 100 released radiographs and 100 reserved as the holdout dataset. The holdout dataset is used exclusively by RSNA to independently evaluate different models. A benchmark of 200 chest radiographic studies with 12 benchmark labels was created and made publicly available https://imaging.rsna.org, with each chest radiograph verified by three radiologists. In addition, an AI-assisted labeling procedure was developed to help radiologists label at scale, minimize unnecessary omissions, and support a semicollaborative environment.
Feature Quality and Adaptability of Medical Foundation Models: A Comparative Evaluation for Radiographic Classification and Segmentation
Nov 12, 2025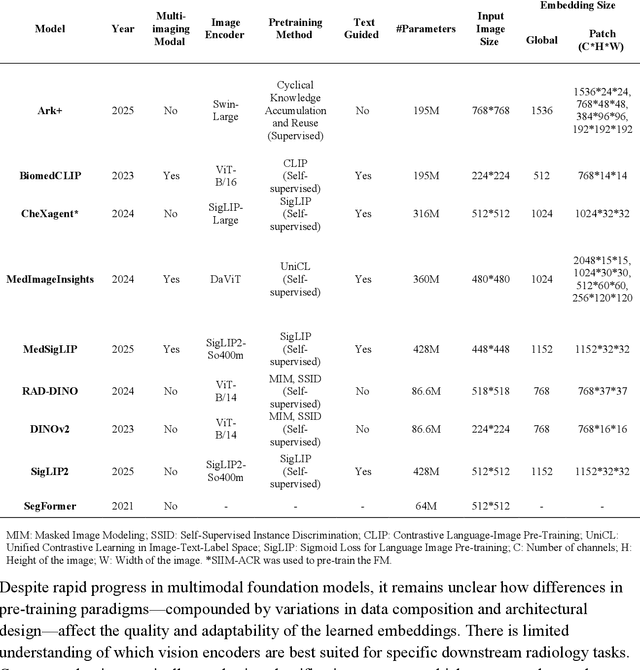
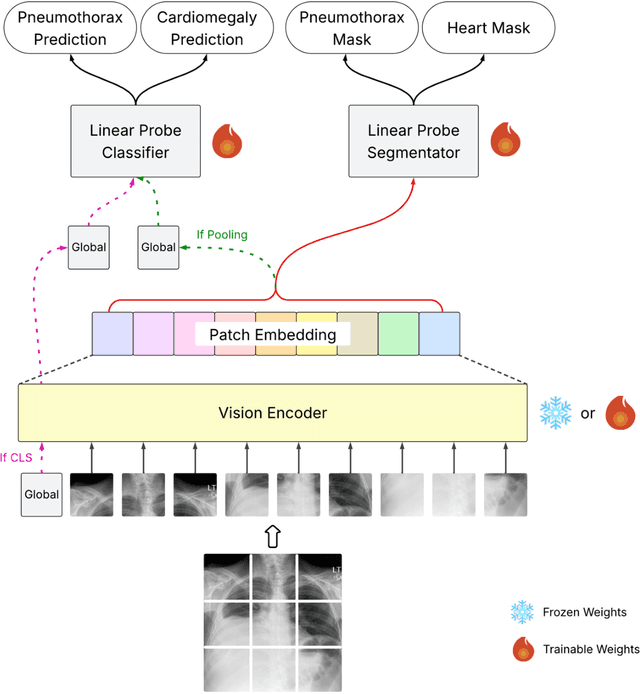
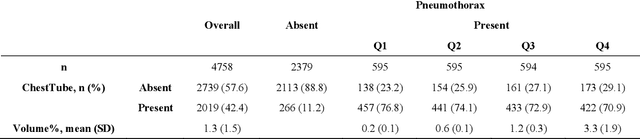
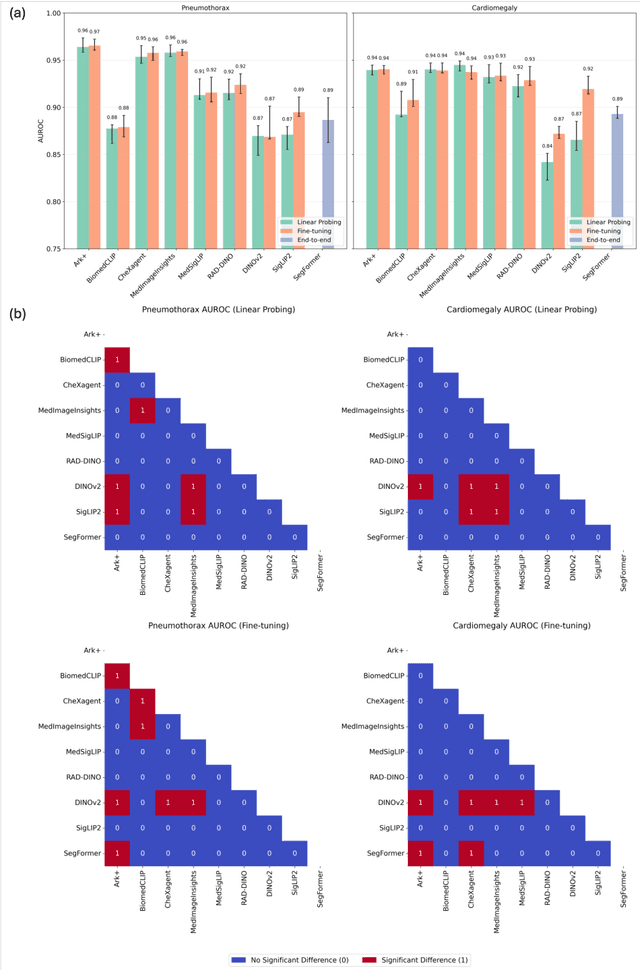
Abstract:Foundation models (FMs) promise to generalize medical imaging, but their effectiveness varies. It remains unclear how pre-training domain (medical vs. general), paradigm (e.g., text-guided), and architecture influence embedding quality, hindering the selection of optimal encoders for specific radiology tasks. To address this, we evaluate vision encoders from eight medical and general-domain FMs for chest X-ray analysis. We benchmark classification (pneumothorax, cardiomegaly) and segmentation (pneumothorax, cardiac boundary) using linear probing and fine-tuning. Our results show that domain-specific pre-training provides a significant advantage; medical FMs consistently outperformed general-domain models in linear probing, establishing superior initial feature quality. However, feature utility is highly task-dependent. Pre-trained embeddings were strong for global classification and segmenting salient anatomy (e.g., heart). In contrast, for segmenting complex, subtle pathologies (e.g., pneumothorax), all FMs performed poorly without significant fine-tuning, revealing a critical gap in localizing subtle disease. Subgroup analysis showed FMs use confounding shortcuts (e.g., chest tubes for pneumothorax) for classification, a strategy that fails for precise segmentation. We also found that expensive text-image alignment is not a prerequisite; image-only (RAD-DINO) and label-supervised (Ark+) FMs were among top performers. Notably, a supervised, end-to-end baseline remained highly competitive, matching or exceeding the best FMs on segmentation tasks. These findings show that while medical pre-training is beneficial, architectural choices (e.g., multi-scale) are critical, and pre-trained features are not universally effective, especially for complex localization tasks where supervised models remain a strong alternative.
Evaluating Vision Language Models (VLMs) for Radiology: A Comprehensive Analysis
Apr 22, 2025



Abstract:Foundation models, trained on vast amounts of data using self-supervised techniques, have emerged as a promising frontier for advancing artificial intelligence (AI) applications in medicine. This study evaluates three different vision-language foundation models (RAD-DINO, CheXagent, and BiomedCLIP) on their ability to capture fine-grained imaging features for radiology tasks. The models were assessed across classification, segmentation, and regression tasks for pneumothorax and cardiomegaly on chest radiographs. Self-supervised RAD-DINO consistently excelled in segmentation tasks, while text-supervised CheXagent demonstrated superior classification performance. BiomedCLIP showed inconsistent performance across tasks. A custom segmentation model that integrates global and local features substantially improved performance for all foundation models, particularly for challenging pneumothorax segmentation. The findings highlight that pre-training methodology significantly influences model performance on specific downstream tasks. For fine-grained segmentation tasks, models trained without text supervision performed better, while text-supervised models offered advantages in classification and interpretability. These insights provide guidance for selecting foundation models based on specific clinical applications in radiology.
A Multi-Modal AI System for Screening Mammography: Integrating 2D and 3D Imaging to Improve Breast Cancer Detection in a Prospective Clinical Study
Apr 08, 2025
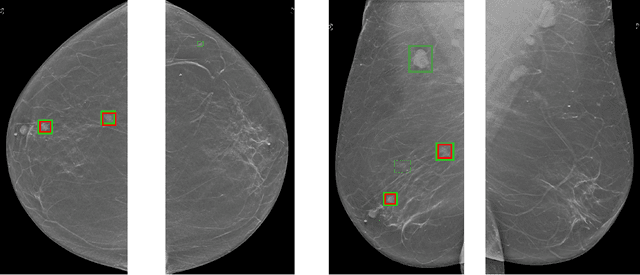
Abstract:Although digital breast tomosynthesis (DBT) improves diagnostic performance over full-field digital mammography (FFDM), false-positive recalls remain a concern in breast cancer screening. We developed a multi-modal artificial intelligence system integrating FFDM, synthetic mammography, and DBT to provide breast-level predictions and bounding-box localizations of suspicious findings. Our AI system, trained on approximately 500,000 mammography exams, achieved 0.945 AUROC on an internal test set. It demonstrated capacity to reduce recalls by 31.7% and radiologist workload by 43.8% while maintaining 100% sensitivity, underscoring its potential to improve clinical workflows. External validation confirmed strong generalizability, reducing the gap to a perfect AUROC by 35.31%-69.14% relative to strong baselines. In prospective deployment across 18 sites, the system reduced recall rates for low-risk cases. An improved version, trained on over 750,000 exams with additional labels, further reduced the gap by 18.86%-56.62% across large external datasets. Overall, these results underscore the importance of utilizing all available imaging modalities, demonstrate the potential for clinical impact, and indicate feasibility of further reduction of the test error with increased training set when using large-capacity neural networks.
Subgroup Performance of a Commercial Digital Breast Tomosynthesis Model for Breast Cancer Detection
Mar 17, 2025Abstract:While research has established the potential of AI models for mammography to improve breast cancer screening outcomes, there have not been any detailed subgroup evaluations performed to assess the strengths and weaknesses of commercial models for digital breast tomosynthesis (DBT) imaging. This study presents a granular evaluation of the Lunit INSIGHT DBT model on a large retrospective cohort of 163,449 screening mammography exams from the Emory Breast Imaging Dataset (EMBED). Model performance was evaluated in a binary context with various negative exam types (162,081 exams) compared against screen detected cancers (1,368 exams) as the positive class. The analysis was stratified across demographic, imaging, and pathologic subgroups to identify potential disparities. The model achieved an overall AUC of 0.91 (95% CI: 0.90-0.92) with a precision of 0.08 (95% CI: 0.08-0.08), and a recall of 0.73 (95% CI: 0.71-0.76). Performance was found to be robust across demographics, but cases with non-invasive cancers (AUC: 0.85, 95% CI: 0.83-0.87), calcifications (AUC: 0.80, 95% CI: 0.78-0.82), and dense breast tissue (AUC: 0.90, 95% CI: 0.88-0.91) were associated with significantly lower performance compared to other groups. These results highlight the need for detailed evaluation of model characteristics and vigilance in considering adoption of new tools for clinical deployment.
Novel AI-Based Quantification of Breast Arterial Calcification to Predict Cardiovascular Risk
Mar 17, 2025Abstract:Women are underdiagnosed and undertreated for cardiovascular disease. Automatic quantification of breast arterial calcification on screening mammography can identify women at risk for cardiovascular disease and enable earlier treatment and management of disease. In this retrospective study of 116,135 women from two healthcare systems, a transformer-based neural network quantified BAC severity (no BAC, mild, moderate, and severe) on screening mammograms. Outcomes included major adverse cardiovascular events (MACE) and all-cause mortality. BAC severity was independently associated with MACE after adjusting for cardiovascular risk factors, with increasing hazard ratios from mild (HR 1.18-1.22), moderate (HR 1.38-1.47), to severe BAC (HR 2.03-2.22) across datasets (all p<0.001). This association remained significant across all age groups, with even mild BAC indicating increased risk in women under 50. BAC remained an independent predictor when analyzed alongside ASCVD risk scores, showing significant associations with myocardial infarction, stroke, heart failure, and mortality (all p<0.005). Automated BAC quantification enables opportunistic cardiovascular risk assessment during routine mammography without additional radiation or cost. This approach provides value beyond traditional risk factors, particularly in younger women, offering potential for early CVD risk stratification in the millions of women undergoing annual mammography.
Emory Knee Radiograph (MRKR) Dataset
Oct 30, 2024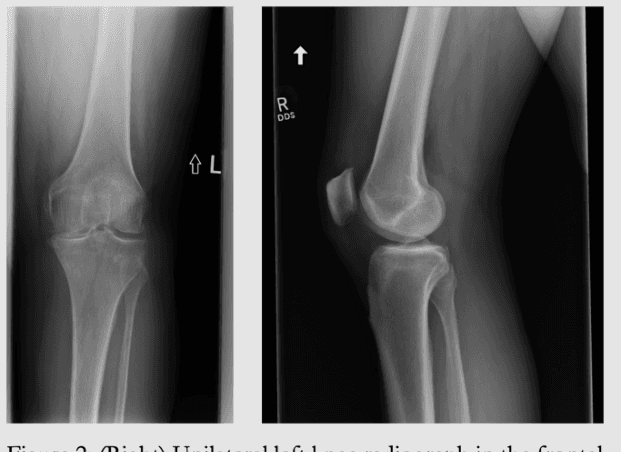
Abstract:The Emory Knee Radiograph (MRKR) dataset is a large, demographically diverse collection of 503,261 knee radiographs from 83,011 patients, 40% of which are African American. This dataset provides imaging data in DICOM format along with detailed clinical information, including patient-reported pain scores, diagnostic codes, and procedural codes, which are not commonly available in similar datasets. The MRKR dataset also features imaging metadata such as image laterality, view type, and presence of hardware, enhancing its value for research and model development. MRKR addresses significant gaps in existing datasets by offering a more representative sample for studying osteoarthritis and related outcomes, particularly among minority populations, thereby providing a valuable resource for clinicians and researchers.
Benchmarking bias: Expanding clinical AI model card to incorporate bias reporting of social and non-social factors
Nov 21, 2023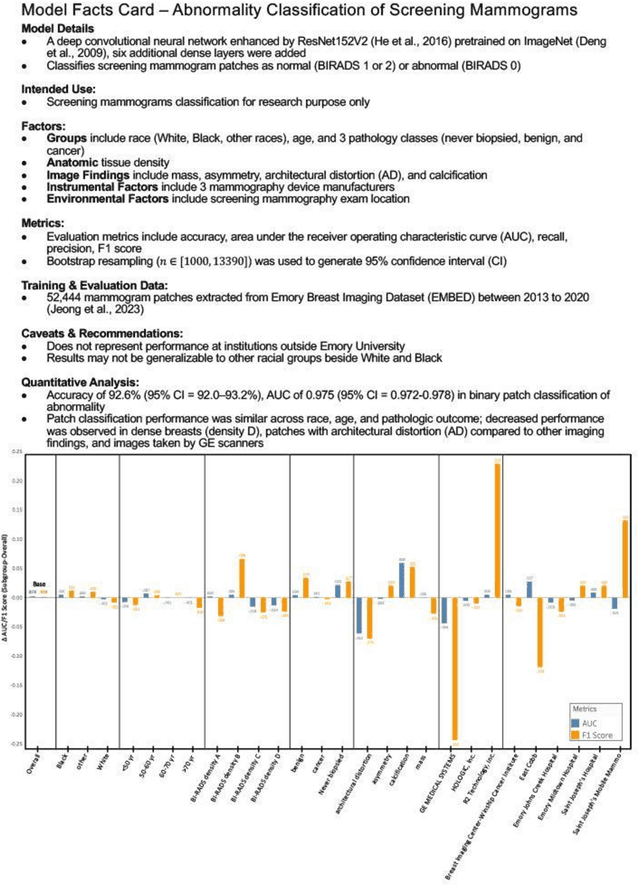
Abstract:Clinical AI model reporting cards should be expanded to incorporate a broad bias reporting of both social and non-social factors. Non-social factors consider the role of other factors, such as disease dependent, anatomic, or instrument factors on AI model bias, which are essential to ensure safe deployment.
Performance Gaps of Artificial Intelligence Models Screening Mammography -- Towards Fair and Interpretable Models
May 08, 2023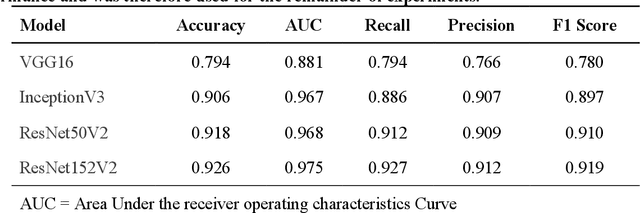
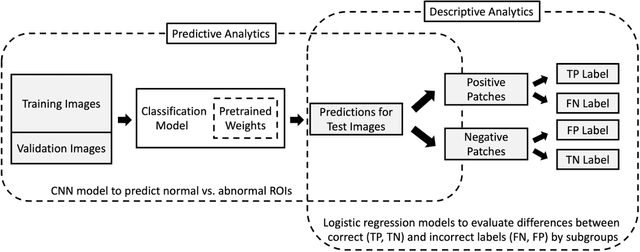
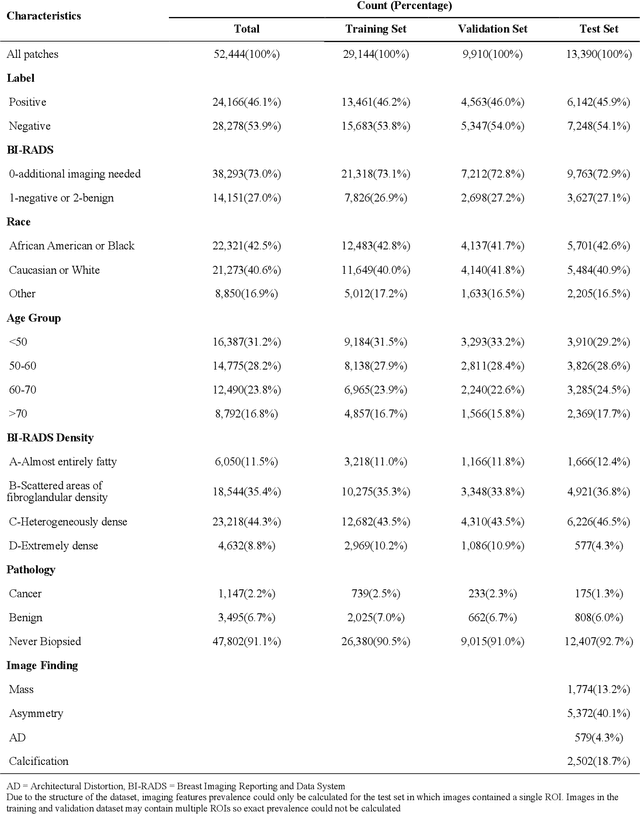
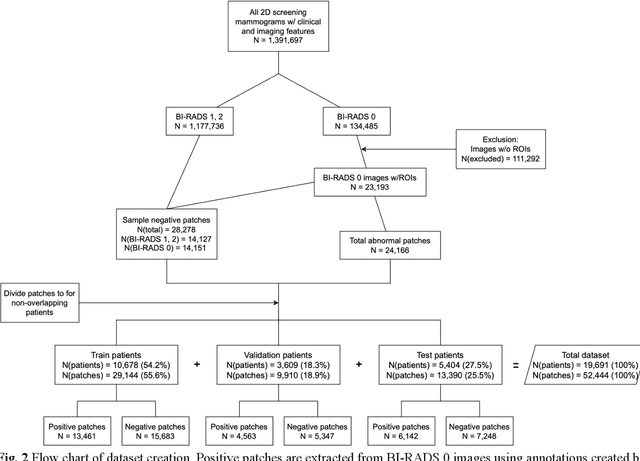
Abstract:Purpose: To analyze the demographic and imaging characteristics associated with increased risk of failure for abnormality classification in screening mammograms. Materials and Methods: This retrospective study used data from the Emory BrEast Imaging Dataset (EMBED) which includes mammograms from 115,931 patients imaged at Emory University Healthcare between 2013 to 2020. Clinical and imaging data includes Breast Imaging Reporting and Data System (BI-RADS) assessment, region of interest coordinates for abnormalities, imaging features, pathologic outcomes, and patient demographics. Multiple deep learning models were developed to distinguish between patches of abnormal tissue and randomly selected patches of normal tissue from the screening mammograms. We assessed model performance overall and within subgroups defined by age, race, pathologic outcome, and imaging characteristics to evaluate reasons for misclassifications. Results: On a test set size of 5,810 studies (13,390 patches), a ResNet152V2 model trained to classify normal versus abnormal tissue patches achieved an accuracy of 92.6% (95% CI = 92.0-93.2%), and area under the receiver operative characteristics curve 0.975 (95% CI = 0.972-0.978). Imaging characteristics associated with higher misclassifications of images include higher tissue densities (risk ratio [RR]=1.649; p=.010, BI-RADS density C and RR=2.026; p=.003, BI-RADS density D), and presence of architectural distortion (RR=1.026; p<.001). Conclusion: Even though deep learning models for abnormality classification can perform well in screening mammography, we demonstrate certain imaging features that result in worse model performance. This is the first such work to systematically evaluate breast abnormality classification by various subgroups and better-informed developers and end-users of population subgroups which are likely to experience biased model performance.
Early Diagnosis of Chronic Obstructive Pulmonary Disease from Chest X-Rays using Transfer Learning and Fusion Strategies
Nov 13, 2022Abstract:Chronic obstructive pulmonary disease (COPD) is one of the most common chronic illnesses in the world and the third leading cause of mortality worldwide. It is often underdiagnosed or not diagnosed until later in the disease course. Spirometry tests are the gold standard for diagnosing COPD but can be difficult to obtain, especially in resource-poor countries. Chest X-rays (CXRs), however, are readily available and may serve as a screening tool to identify patients with COPD who should undergo further testing. Currently, no research applies deep learning (DL) algorithms that use large multi-site and multi-modal data to detect COPD patients and evaluate fairness across demographic groups. We use three CXR datasets in our study, CheXpert to pre-train models, MIMIC-CXR to develop, and Emory-CXR to validate our models. The CXRs from patients in the early stage of COPD and not on mechanical ventilation are selected for model training and validation. We visualize the Grad-CAM heatmaps of the true positive cases on the base model for both MIMIC-CXR and Emory-CXR test datasets. We further propose two fusion schemes, (1) model-level fusion, including bagging and stacking methods using MIMIC-CXR, and (2) data-level fusion, including multi-site data using MIMIC-CXR and Emory-CXR, and multi-modal using MIMIC-CXRs and MIMIC-IV EHR, to improve the overall model performance. Fairness analysis is performed to evaluate if the fusion schemes have a discrepancy in the performance among different demographic groups. The results demonstrate that DL models can detect COPD using CXRs, which can facilitate early screening, especially in low-resource regions where CXRs are more accessible than spirometry. The multi-site data fusion scheme could improve the model generalizability on the Emory-CXR test data. Further studies on using CXR or other modalities to predict COPD ought to be in future work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge