Leo Anthony Celi
MIT
Uncertainty Makes It Stable: Curiosity-Driven Quantized Mixture-of-Experts
Nov 19, 2025Abstract:Deploying deep neural networks on resource-constrained devices faces two critical challenges: maintaining accuracy under aggressive quantization while ensuring predictable inference latency. We present a curiosity-driven quantized Mixture-of-Experts framework that addresses both through Bayesian epistemic uncertainty-based routing across heterogeneous experts (BitNet ternary, 1-16 bit BitLinear, post-training quantization). Evaluated on audio classification benchmarks (ESC-50, Quinn, UrbanSound8K), our 4-bit quantization maintains 99.9 percent of 16-bit accuracy (0.858 vs 0.859 F1) with 4x compression and 41 percent energy savings versus 8-bit. Crucially, curiosity-driven routing reduces MoE latency variance by 82 percent (p = 0.008, Levene's test) from 230 ms to 29 ms standard deviation, enabling stable inference for battery-constrained devices. Statistical analysis confirms 4-bit/8-bit achieve practical equivalence with full precision (p > 0.05), while MoE architectures introduce 11 percent latency overhead (p < 0.001) without accuracy gains. At scale, deployment emissions dominate training by 10000x for models serving more than 1,000 inferences, making inference efficiency critical. Our information-theoretic routing demonstrates that adaptive quantization yields accurate (0.858 F1, 1.2M params), energy-efficient (3.87 F1/mJ), and predictable edge models, with simple 4-bit quantized architectures outperforming complex MoE for most deployments.
Algorithms Trained on Normal Chest X-rays Can Predict Health Insurance Types
Nov 17, 2025Abstract:Artificial intelligence is revealing what medicine never intended to encode. Deep vision models, trained on chest X-rays, can now detect not only disease but also invisible traces of social inequality. In this study, we show that state-of-the-art architectures (DenseNet121, SwinV2-B, MedMamba) can predict a patient's health insurance type, a strong proxy for socioeconomic status, from normal chest X-rays with significant accuracy (AUC around 0.67 on MIMIC-CXR-JPG, 0.68 on CheXpert). The signal persists even when age, race, and sex are controlled for, and remains detectable when the model is trained exclusively on a single racial group. Patch-based occlusion reveals that the signal is diffuse rather than localized, embedded in the upper and mid-thoracic regions. This suggests that deep networks may be internalizing subtle traces of clinical environments, equipment differences, or care pathways; learning socioeconomic segregation itself. These findings challenge the assumption that medical images are neutral biological data. By uncovering how models perceive and exploit these hidden social signatures, this work reframes fairness in medical AI: the goal is no longer only to balance datasets or adjust thresholds, but to interrogate and disentangle the social fingerprints embedded in clinical data itself.
CXR-LT 2024: A MICCAI challenge on long-tailed, multi-label, and zero-shot disease classification from chest X-ray
Jun 09, 2025Abstract:The CXR-LT series is a community-driven initiative designed to enhance lung disease classification using chest X-rays (CXR). It tackles challenges in open long-tailed lung disease classification and enhances the measurability of state-of-the-art techniques. The first event, CXR-LT 2023, aimed to achieve these goals by providing high-quality benchmark CXR data for model development and conducting comprehensive evaluations to identify ongoing issues impacting lung disease classification performance. Building on the success of CXR-LT 2023, the CXR-LT 2024 expands the dataset to 377,110 chest X-rays (CXRs) and 45 disease labels, including 19 new rare disease findings. It also introduces a new focus on zero-shot learning to address limitations identified in the previous event. Specifically, CXR-LT 2024 features three tasks: (i) long-tailed classification on a large, noisy test set, (ii) long-tailed classification on a manually annotated "gold standard" subset, and (iii) zero-shot generalization to five previously unseen disease findings. This paper provides an overview of CXR-LT 2024, detailing the data curation process and consolidating state-of-the-art solutions, including the use of multimodal models for rare disease detection, advanced generative approaches to handle noisy labels, and zero-shot learning strategies for unseen diseases. Additionally, the expanded dataset enhances disease coverage to better represent real-world clinical settings, offering a valuable resource for future research. By synthesizing the insights and innovations of participating teams, we aim to advance the development of clinically realistic and generalizable diagnostic models for chest radiography.
Performance Gains of LLMs With Humans in a World of LLMs Versus Humans
May 13, 2025



Abstract:Currently, a considerable research effort is devoted to comparing LLMs to a group of human experts, where the term "expert" is often ill-defined or variable, at best, in a state of constantly updating LLM releases. Without proper safeguards in place, LLMs will threaten to cause harm to the established structure of safe delivery of patient care which has been carefully developed throughout history to keep the safety of the patient at the forefront. A key driver of LLM innovation is founded on community research efforts which, if continuing to operate under "humans versus LLMs" principles, will expedite this trend. Therefore, research efforts moving forward must focus on effectively characterizing the safe use of LLMs in clinical settings that persist across the rapid development of novel LLM models. In this communication, we demonstrate that rather than comparing LLMs to humans, there is a need to develop strategies enabling efficient work of humans with LLMs in an almost symbiotic manner.
BRIDGE: Benchmarking Large Language Models for Understanding Real-world Clinical Practice Text
May 01, 2025



Abstract:Large language models (LLMs) hold great promise for medical applications and are evolving rapidly, with new models being released at an accelerated pace. However, current evaluations of LLMs in clinical contexts remain limited. Most existing benchmarks rely on medical exam-style questions or PubMed-derived text, failing to capture the complexity of real-world electronic health record (EHR) data. Others focus narrowly on specific application scenarios, limiting their generalizability across broader clinical use. To address this gap, we present BRIDGE, a comprehensive multilingual benchmark comprising 87 tasks sourced from real-world clinical data sources across nine languages. We systematically evaluated 52 state-of-the-art LLMs (including DeepSeek-R1, GPT-4o, Gemini, and Llama 4) under various inference strategies. With a total of 13,572 experiments, our results reveal substantial performance variation across model sizes, languages, natural language processing tasks, and clinical specialties. Notably, we demonstrate that open-source LLMs can achieve performance comparable to proprietary models, while medically fine-tuned LLMs based on older architectures often underperform versus updated general-purpose models. The BRIDGE and its corresponding leaderboard serve as a foundational resource and a unique reference for the development and evaluation of new LLMs in real-world clinical text understanding. The BRIDGE leaderboard: https://huggingface.co/spaces/YLab-Open/BRIDGE-Medical-Leaderboard
An Algorithmic Approach for Causal Health Equity: A Look at Race Differentials in Intensive Care Unit (ICU) Outcomes
Jan 09, 2025
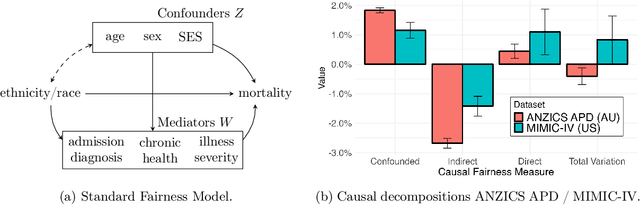
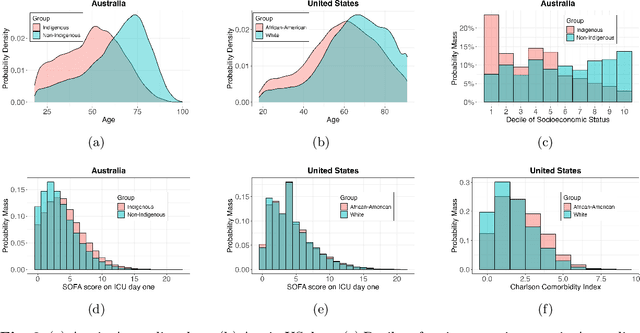

Abstract:The new era of large-scale data collection and analysis presents an opportunity for diagnosing and understanding the causes of health inequities. In this study, we describe a framework for systematically analyzing health disparities using causal inference. The framework is illustrated by investigating racial and ethnic disparities in intensive care unit (ICU) outcome between majority and minority groups in Australia (Indigenous vs. Non-Indigenous) and the United States (African-American vs. White). We demonstrate that commonly used statistical measures for quantifying inequity are insufficient, and focus on attributing the observed disparity to the causal mechanisms that generate it. We find that minority patients are younger at admission, have worse chronic health, are more likely to be admitted for urgent and non-elective reasons, and have higher illness severity. At the same time, however, we find a protective direct effect of belonging to a minority group, with minority patients showing improved survival compared to their majority counterparts, with all other variables kept equal. We demonstrate that this protective effect is related to the increased probability of being admitted to ICU, with minority patients having an increased risk of ICU admission. We also find that minority patients, while showing improved survival, are more likely to be readmitted to ICU. Thus, due to worse access to primary health care, minority patients are more likely to end up in ICU for preventable conditions, causing a reduction in the mortality rates and creating an effect that appears to be protective. Since the baseline risk of ICU admission may serve as proxy for lack of access to primary care, we developed the Indigenous Intensive Care Equity (IICE) Radar, a monitoring system for tracking the over-utilization of ICU resources by the Indigenous population of Australia across geographical areas.
Multi-OphthaLingua: A Multilingual Benchmark for Assessing and Debiasing LLM Ophthalmological QA in LMICs
Dec 18, 2024



Abstract:Current ophthalmology clinical workflows are plagued by over-referrals, long waits, and complex and heterogeneous medical records. Large language models (LLMs) present a promising solution to automate various procedures such as triaging, preliminary tests like visual acuity assessment, and report summaries. However, LLMs have demonstrated significantly varied performance across different languages in natural language question-answering tasks, potentially exacerbating healthcare disparities in Low and Middle-Income Countries (LMICs). This study introduces the first multilingual ophthalmological question-answering benchmark with manually curated questions parallel across languages, allowing for direct cross-lingual comparisons. Our evaluation of 6 popular LLMs across 7 different languages reveals substantial bias across different languages, highlighting risks for clinical deployment of LLMs in LMICs. Existing debiasing methods such as Translation Chain-of-Thought or Retrieval-augmented generation (RAG) by themselves fall short of closing this performance gap, often failing to improve performance across all languages and lacking specificity for the medical domain. To address this issue, We propose CLARA (Cross-Lingual Reflective Agentic system), a novel inference time de-biasing method leveraging retrieval augmented generation and self-verification. Our approach not only improves performance across all languages but also significantly reduces the multilingual bias gap, facilitating equitable LLM application across the globe.
MedDec: A Dataset for Extracting Medical Decisions from Discharge Summaries
Aug 23, 2024
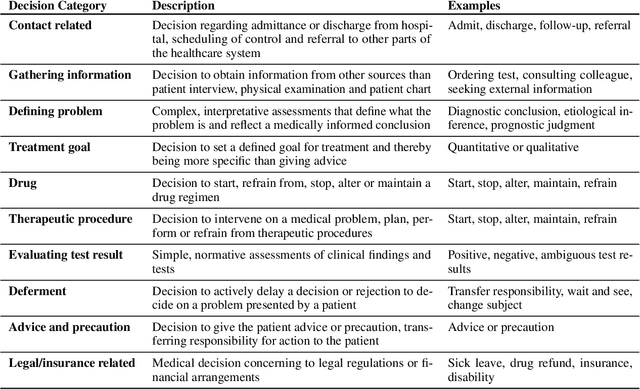
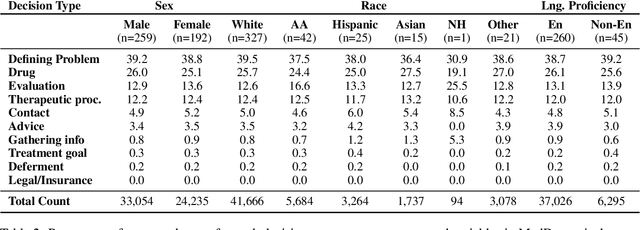
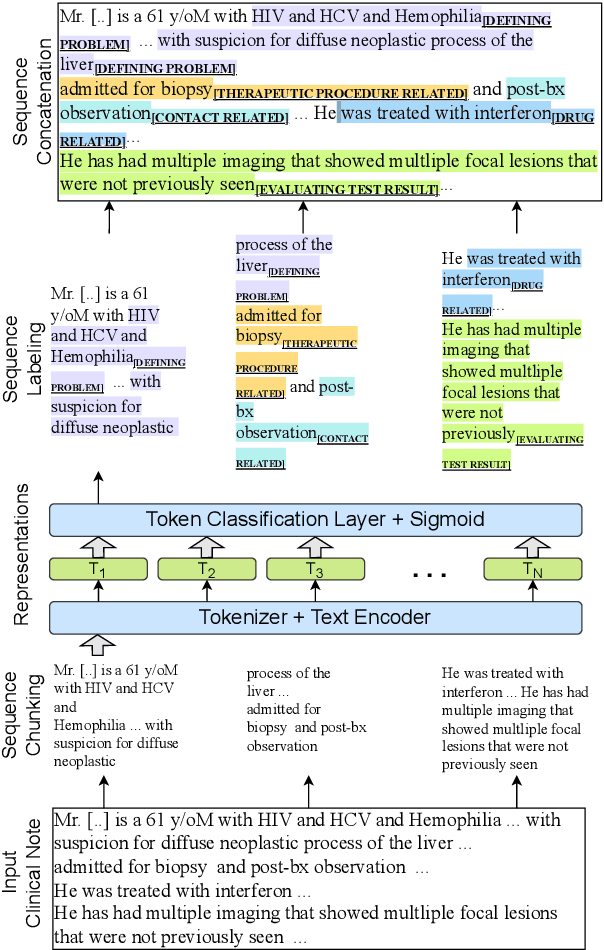
Abstract:Medical decisions directly impact individuals' health and well-being. Extracting decision spans from clinical notes plays a crucial role in understanding medical decision-making processes. In this paper, we develop a new dataset called "MedDec", which contains clinical notes of eleven different phenotypes (diseases) annotated by ten types of medical decisions. We introduce the task of medical decision extraction, aiming to jointly extract and classify different types of medical decisions within clinical notes. We provide a comprehensive analysis of the dataset, develop a span detection model as a baseline for this task, evaluate recent span detection approaches, and employ a few metrics to measure the complexity of data samples. Our findings shed light on the complexities inherent in clinical decision extraction and enable future work in this area of research. The dataset and code are available through https://github.com/CLU-UML/MedDec.
Language Models are Surprisingly Fragile to Drug Names in Biomedical Benchmarks
Jun 17, 2024Abstract:Medical knowledge is context-dependent and requires consistent reasoning across various natural language expressions of semantically equivalent phrases. This is particularly crucial for drug names, where patients often use brand names like Advil or Tylenol instead of their generic equivalents. To study this, we create a new robustness dataset, RABBITS, to evaluate performance differences on medical benchmarks after swapping brand and generic drug names using physician expert annotations. We assess both open-source and API-based LLMs on MedQA and MedMCQA, revealing a consistent performance drop ranging from 1-10\%. Furthermore, we identify a potential source of this fragility as the contamination of test data in widely used pre-training datasets. All code is accessible at https://github.com/BittermanLab/RABBITS, and a HuggingFace leaderboard is available at https://huggingface.co/spaces/AIM-Harvard/rabbits-leaderboard.
Multimodal Deep Learning for Low-Resource Settings: A Vector Embedding Alignment Approach for Healthcare Applications
Jun 02, 2024Abstract:Large-scale multi-modal deep learning models have revolutionized domains such as healthcare, highlighting the importance of computational power. However, in resource-constrained regions like Low and Middle-Income Countries (LMICs), limited access to GPUs and data poses significant challenges, often leaving CPUs as the sole resource. To address this, we advocate for leveraging vector embeddings to enable flexible and efficient computational methodologies, democratizing multimodal deep learning across diverse contexts. Our paper investigates the efficiency and effectiveness of using vector embeddings from single-modal foundation models and multi-modal Vision-Language Models (VLMs) for multimodal deep learning in low-resource environments, particularly in healthcare. Additionally, we propose a simple yet effective inference-time method to enhance performance by aligning image-text embeddings. Comparing these approaches with traditional methods, we assess their impact on computational efficiency and model performance using metrics like accuracy, F1-score, inference time, training time, and memory usage across three medical modalities: BRSET (ophthalmology), HAM10000 (dermatology), and SatelliteBench (public health). Our findings show that embeddings reduce computational demands without compromising model performance. Furthermore, our alignment method improves performance in medical tasks. This research promotes sustainable AI practices by optimizing resources in constrained environments, highlighting the potential of embedding-based approaches for efficient multimodal learning. Vector embeddings democratize multimodal deep learning in LMICs, particularly in healthcare, enhancing AI adaptability in varied use cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge