Jack Gallifant
Massachusetts Institute of Technology
KScope: A Framework for Characterizing the Knowledge Status of Language Models
Jun 09, 2025Abstract:Characterizing a large language model's (LLM's) knowledge of a given question is challenging. As a result, prior work has primarily examined LLM behavior under knowledge conflicts, where the model's internal parametric memory contradicts information in the external context. However, this does not fully reflect how well the model knows the answer to the question. In this paper, we first introduce a taxonomy of five knowledge statuses based on the consistency and correctness of LLM knowledge modes. We then propose KScope, a hierarchical framework of statistical tests that progressively refines hypotheses about knowledge modes and characterizes LLM knowledge into one of these five statuses. We apply KScope to nine LLMs across four datasets and systematically establish: (1) Supporting context narrows knowledge gaps across models. (2) Context features related to difficulty, relevance, and familiarity drive successful knowledge updates. (3) LLMs exhibit similar feature preferences when partially correct or conflicted, but diverge sharply when consistently wrong. (4) Context summarization constrained by our feature analysis, together with enhanced credibility, further improves update effectiveness and generalizes across LLMs.
MedBrowseComp: Benchmarking Medical Deep Research and Computer Use
May 20, 2025Abstract:Large language models (LLMs) are increasingly envisioned as decision-support tools in clinical practice, yet safe clinical reasoning demands integrating heterogeneous knowledge bases -- trials, primary studies, regulatory documents, and cost data -- under strict accuracy constraints. Existing evaluations often rely on synthetic prompts, reduce the task to single-hop factoid queries, or conflate reasoning with open-ended generation, leaving their real-world utility unclear. To close this gap, we present MedBrowseComp, the first benchmark that systematically tests an agent's ability to reliably retrieve and synthesize multi-hop medical facts from live, domain-specific knowledge bases. MedBrowseComp contains more than 1,000 human-curated questions that mirror clinical scenarios where practitioners must reconcile fragmented or conflicting information to reach an up-to-date conclusion. Applying MedBrowseComp to frontier agentic systems reveals performance shortfalls as low as ten percent, exposing a critical gap between current LLM capabilities and the rigor demanded in clinical settings. MedBrowseComp therefore offers a clear testbed for reliable medical information seeking and sets concrete goals for future model and toolchain upgrades. You can visit our project page at: https://moreirap12.github.io/mbc-browse-app/
Sparse Autoencoder Features for Classifications and Transferability
Feb 17, 2025Abstract:Sparse Autoencoders (SAEs) provide potentials for uncovering structured, human-interpretable representations in Large Language Models (LLMs), making them a crucial tool for transparent and controllable AI systems. We systematically analyze SAE for interpretable feature extraction from LLMs in safety-critical classification tasks. Our framework evaluates (1) model-layer selection and scaling properties, (2) SAE architectural configurations, including width and pooling strategies, and (3) the effect of binarizing continuous SAE activations. SAE-derived features achieve macro F1 > 0.8, outperforming hidden-state and BoW baselines while demonstrating cross-model transfer from Gemma 2 2B to 9B-IT models. These features generalize in a zero-shot manner to cross-lingual toxicity detection and visual classification tasks. Our analysis highlights the significant impact of pooling strategies and binarization thresholds, showing that binarization offers an efficient alternative to traditional feature selection while maintaining or improving performance. These findings establish new best practices for SAE-based interpretability and enable scalable, transparent deployment of LLMs in real-world applications. Full repo: https://github.com/shan23chen/MOSAIC.
Representation Learning of Lab Values via Masked AutoEncoder
Jan 05, 2025



Abstract:Accurate imputation of missing laboratory values in electronic health records (EHRs) is critical to enable robust clinical predictions and reduce biases in AI systems in healthcare. Existing methods, such as variational autoencoders (VAEs) and decision tree-based approaches such as XGBoost, struggle to model the complex temporal and contextual dependencies in EHR data, mainly in underrepresented groups. In this work, we propose Lab-MAE, a novel transformer-based masked autoencoder framework that leverages self-supervised learning for the imputation of continuous sequential lab values. Lab-MAE introduces a structured encoding scheme that jointly models laboratory test values and their corresponding timestamps, enabling explicit capturing temporal dependencies. Empirical evaluation on the MIMIC-IV dataset demonstrates that Lab-MAE significantly outperforms the state-of-the-art baselines such as XGBoost across multiple metrics, including root mean square error (RMSE), R-squared (R2), and Wasserstein distance (WD). Notably, Lab-MAE achieves equitable performance across demographic groups of patients, advancing fairness in clinical predictions. We further investigate the role of follow-up laboratory values as potential shortcut features, revealing Lab-MAE's robustness in scenarios where such data is unavailable. The findings suggest that our transformer-based architecture, adapted to the characteristics of the EHR data, offers a foundation model for more accurate and fair clinical imputation models. In addition, we measure and compare the carbon footprint of Lab-MAE with the baseline XGBoost model, highlighting its environmental requirements.
Multi-OphthaLingua: A Multilingual Benchmark for Assessing and Debiasing LLM Ophthalmological QA in LMICs
Dec 18, 2024



Abstract:Current ophthalmology clinical workflows are plagued by over-referrals, long waits, and complex and heterogeneous medical records. Large language models (LLMs) present a promising solution to automate various procedures such as triaging, preliminary tests like visual acuity assessment, and report summaries. However, LLMs have demonstrated significantly varied performance across different languages in natural language question-answering tasks, potentially exacerbating healthcare disparities in Low and Middle-Income Countries (LMICs). This study introduces the first multilingual ophthalmological question-answering benchmark with manually curated questions parallel across languages, allowing for direct cross-lingual comparisons. Our evaluation of 6 popular LLMs across 7 different languages reveals substantial bias across different languages, highlighting risks for clinical deployment of LLMs in LMICs. Existing debiasing methods such as Translation Chain-of-Thought or Retrieval-augmented generation (RAG) by themselves fall short of closing this performance gap, often failing to improve performance across all languages and lacking specificity for the medical domain. To address this issue, We propose CLARA (Cross-Lingual Reflective Agentic system), a novel inference time de-biasing method leveraging retrieval augmented generation and self-verification. Our approach not only improves performance across all languages but also significantly reduces the multilingual bias gap, facilitating equitable LLM application across the globe.
The use of large language models to enhance cancer clinical trial educational materials
Dec 02, 2024Abstract:Cancer clinical trials often face challenges in recruitment and engagement due to a lack of participant-facing informational and educational resources. This study investigated the potential of Large Language Models (LLMs), specifically GPT4, in generating patient-friendly educational content from clinical trial informed consent forms. Using data from ClinicalTrials.gov, we employed zero-shot learning for creating trial summaries and one-shot learning for developing multiple-choice questions, evaluating their effectiveness through patient surveys and crowdsourced annotation. Results showed that GPT4-generated summaries were both readable and comprehensive, and may improve patients' understanding and interest in clinical trials. The multiple-choice questions demonstrated high accuracy and agreement with crowdsourced annotators. For both resource types, hallucinations were identified that require ongoing human oversight. The findings demonstrate the potential of LLMs "out-of-the-box" to support the generation of clinical trial education materials with minimal trial-specific engineering, but implementation with a human-in-the-loop is still needed to avoid misinformation risks.
WorldMedQA-V: a multilingual, multimodal medical examination dataset for multimodal language models evaluation
Oct 16, 2024Abstract:Multimodal/vision language models (VLMs) are increasingly being deployed in healthcare settings worldwide, necessitating robust benchmarks to ensure their safety, efficacy, and fairness. Multiple-choice question and answer (QA) datasets derived from national medical examinations have long served as valuable evaluation tools, but existing datasets are largely text-only and available in a limited subset of languages and countries. To address these challenges, we present WorldMedQA-V, an updated multilingual, multimodal benchmarking dataset designed to evaluate VLMs in healthcare. WorldMedQA-V includes 568 labeled multiple-choice QAs paired with 568 medical images from four countries (Brazil, Israel, Japan, and Spain), covering original languages and validated English translations by native clinicians, respectively. Baseline performance for common open- and closed-source models are provided in the local language and English translations, and with and without images provided to the model. The WorldMedQA-V benchmark aims to better match AI systems to the diverse healthcare environments in which they are deployed, fostering more equitable, effective, and representative applications.
Wait, but Tylenol is Acetaminophen... Investigating and Improving Language Models' Ability to Resist Requests for Misinformation
Sep 30, 2024Abstract:Background: Large language models (LLMs) are trained to follow directions, but this introduces a vulnerability to blindly comply with user requests even if they generate wrong information. In medicine, this could accelerate the generation of misinformation that impacts human well-being. Objectives/Methods: We analyzed compliance to requests to generate misleading content about medications in settings where models know the request is illogical. We investigated whether in-context directions and instruction-tuning of LLMs to prioritize logical reasoning over compliance reduced misinformation risk. Results: While all frontier LLMs complied with misinformation requests, both prompt-based and parameter-based approaches can improve the detection of logic flaws in requests and prevent the dissemination of medical misinformation. Conclusion: Shifting LLMs to prioritize logic over compliance could reduce risks of exploitation for medical misinformation.
AIPatient: Simulating Patients with EHRs and LLM Powered Agentic Workflow
Sep 27, 2024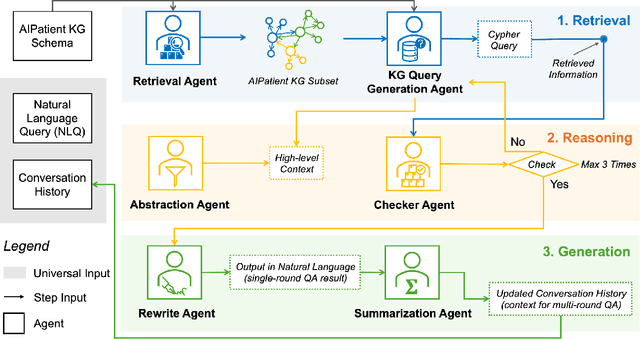
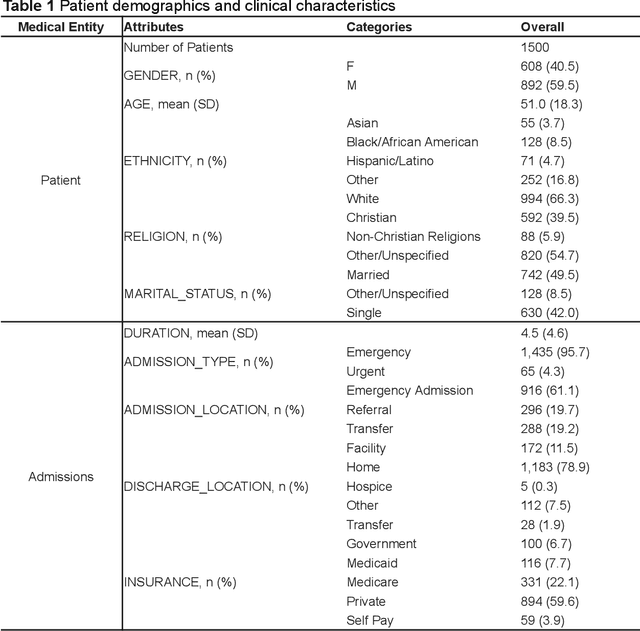
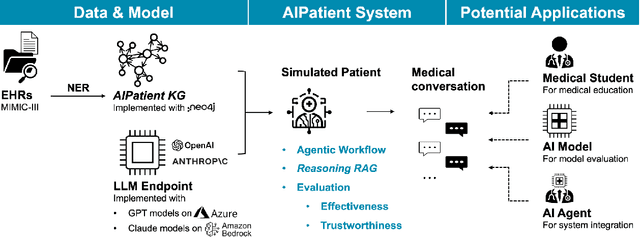
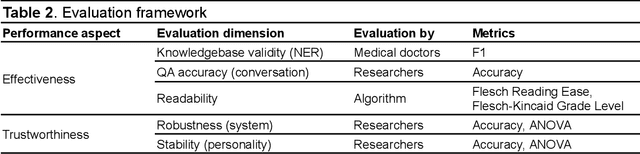
Abstract:Simulated patient systems play a crucial role in modern medical education and research, providing safe, integrative learning environments and enabling clinical decision-making simulations. Large Language Models (LLM) could advance simulated patient systems by replicating medical conditions and patient-doctor interactions with high fidelity and low cost. However, ensuring the effectiveness and trustworthiness of these systems remains a challenge, as they require a large, diverse, and precise patient knowledgebase, along with a robust and stable knowledge diffusion to users. Here, we developed AIPatient, an advanced simulated patient system with AIPatient Knowledge Graph (AIPatient KG) as the input and the Reasoning Retrieval-Augmented Generation (Reasoning RAG) agentic workflow as the generation backbone. AIPatient KG samples data from Electronic Health Records (EHRs) in the Medical Information Mart for Intensive Care (MIMIC)-III database, producing a clinically diverse and relevant cohort of 1,495 patients with high knowledgebase validity (F1 0.89). Reasoning RAG leverages six LLM powered agents spanning tasks including retrieval, KG query generation, abstraction, checker, rewrite, and summarization. This agentic framework reaches an overall accuracy of 94.15% in EHR-based medical Question Answering (QA), outperforming benchmarks that use either no agent or only partial agent integration. Our system also presents high readability (median Flesch Reading Ease 77.23; median Flesch Kincaid Grade 5.6), robustness (ANOVA F-value 0.6126, p<0.1), and stability (ANOVA F-value 0.782, p<0.1). The promising performance of the AIPatient system highlights its potential to support a wide range of applications, including medical education, model evaluation, and system integration.
ACES: Automatic Cohort Extraction System for Event-Stream Datasets
Jun 28, 2024Abstract:Reproducibility remains a significant challenge in machine learning (ML) for healthcare. In this field, datasets, model pipelines, and even task/cohort definitions are often private, leading to a significant barrier in sharing, iterating, and understanding ML results on electronic health record (EHR) datasets. In this paper, we address a significant part of this problem by introducing the Automatic Cohort Extraction System for Event-Stream Datasets (ACES). This tool is designed to simultaneously simplify the development of task/cohorts for ML in healthcare and enable the reproduction of these cohorts, both at an exact level for single datasets and at a conceptual level across datasets. To accomplish this, ACES provides (1) a highly intuitive and expressive configuration language for defining both dataset-specific concepts and dataset-agnostic inclusion/exclusion criteria, and (2) a pipeline to automatically extract patient records that meet these defined criteria from real-world data. ACES can be automatically applied to any dataset in either the Medical Event Data Standard (MEDS) or EventStreamGPT (ESGPT) formats, or to *any* dataset for which the necessary task-specific predicates can be extracted in an event-stream form. ACES has the potential to significantly lower the barrier to entry for defining ML tasks, redefine the way researchers interact with EHR datasets, and significantly improve the state of reproducibility for ML studies in this modality. ACES is available at https://github.com/justin13601/aces.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge