Alistair E. W. Johnson
BioClinical ModernBERT: A State-of-the-Art Long-Context Encoder for Biomedical and Clinical NLP
Jun 12, 2025Abstract:Encoder-based transformer models are central to biomedical and clinical Natural Language Processing (NLP), as their bidirectional self-attention makes them well-suited for efficiently extracting structured information from unstructured text through discriminative tasks. However, encoders have seen slower development compared to decoder models, leading to limited domain adaptation in biomedical and clinical settings. We introduce BioClinical ModernBERT, a domain-adapted encoder that builds on the recent ModernBERT release, incorporating long-context processing and substantial improvements in speed and performance for biomedical and clinical NLP. BioClinical ModernBERT is developed through continued pretraining on the largest biomedical and clinical corpus to date, with over 53.5 billion tokens, and addresses a key limitation of prior clinical encoders by leveraging 20 datasets from diverse institutions, domains, and geographic regions, rather than relying on data from a single source. It outperforms existing biomedical and clinical encoders on four downstream tasks spanning a broad range of use cases. We release both base (150M parameters) and large (396M parameters) versions of BioClinical ModernBERT, along with training checkpoints to support further research.
ACES: Automatic Cohort Extraction System for Event-Stream Datasets
Jun 28, 2024Abstract:Reproducibility remains a significant challenge in machine learning (ML) for healthcare. In this field, datasets, model pipelines, and even task/cohort definitions are often private, leading to a significant barrier in sharing, iterating, and understanding ML results on electronic health record (EHR) datasets. In this paper, we address a significant part of this problem by introducing the Automatic Cohort Extraction System for Event-Stream Datasets (ACES). This tool is designed to simultaneously simplify the development of task/cohorts for ML in healthcare and enable the reproduction of these cohorts, both at an exact level for single datasets and at a conceptual level across datasets. To accomplish this, ACES provides (1) a highly intuitive and expressive configuration language for defining both dataset-specific concepts and dataset-agnostic inclusion/exclusion criteria, and (2) a pipeline to automatically extract patient records that meet these defined criteria from real-world data. ACES can be automatically applied to any dataset in either the Medical Event Data Standard (MEDS) or EventStreamGPT (ESGPT) formats, or to *any* dataset for which the necessary task-specific predicates can be extracted in an event-stream form. ACES has the potential to significantly lower the barrier to entry for defining ML tasks, redefine the way researchers interact with EHR datasets, and significantly improve the state of reproducibility for ML studies in this modality. ACES is available at https://github.com/justin13601/aces.
MIMIC-CXR: A large publicly available database of labeled chest radiographs
Jan 23, 2019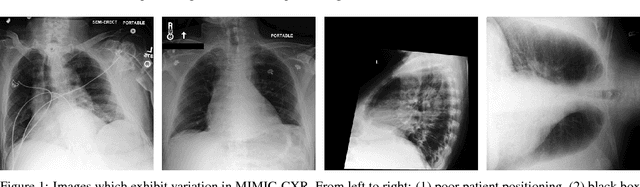
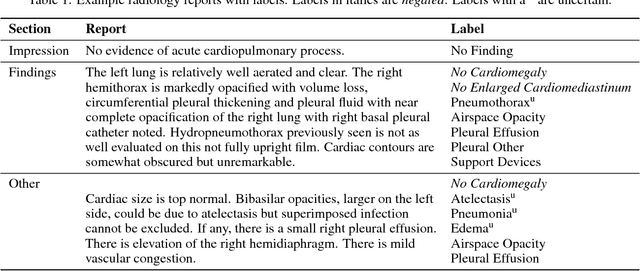
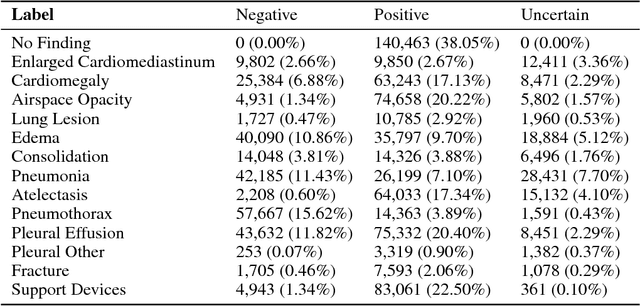
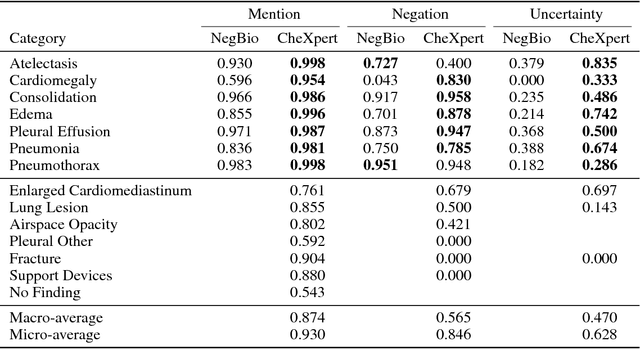
Abstract:Chest radiography is an extremely powerful imaging modality, allowing for a detailed inspection of a patient's thorax, but requiring specialized training for proper interpretation. With the advent of high performance general purpose computer vision algorithms, the accurate automated analysis of chest radiographs is becoming increasingly of interest to researchers. However, a key challenge in the development of these techniques is the lack of sufficient data. Here we describe MIMIC-CXR, a large dataset of 371,920 chest x-rays associated with 227,943 imaging studies sourced from the Beth Israel Deaconess Medical Center between 2011 - 2016. Each imaging study can pertain to one or more images, but most often are associated with two images: a frontal view and a lateral view. Images are provided with 14 labels derived from a natural language processing tool applied to the corresponding free-text radiology reports. All images have been de-identified to protect patient privacy. The dataset is made freely available to facilitate and encourage a wide range of research in medical computer vision.
Generalizability of predictive models for intensive care unit patients
Dec 06, 2018
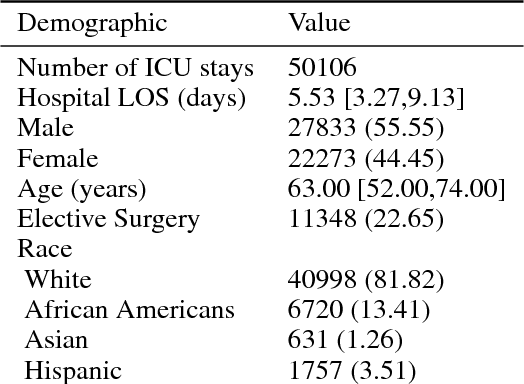
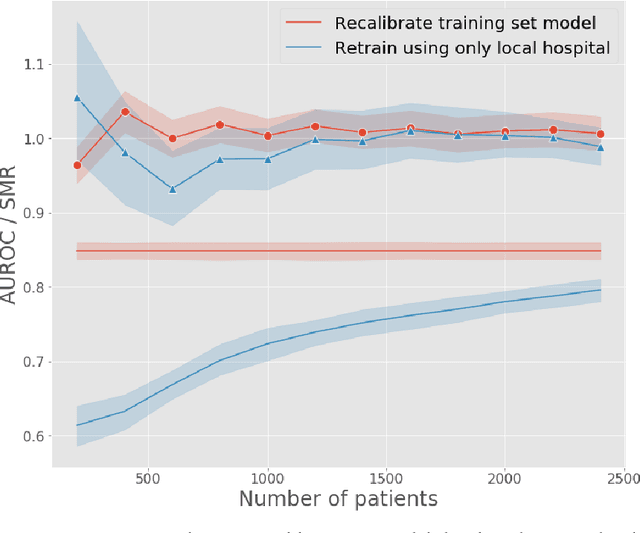
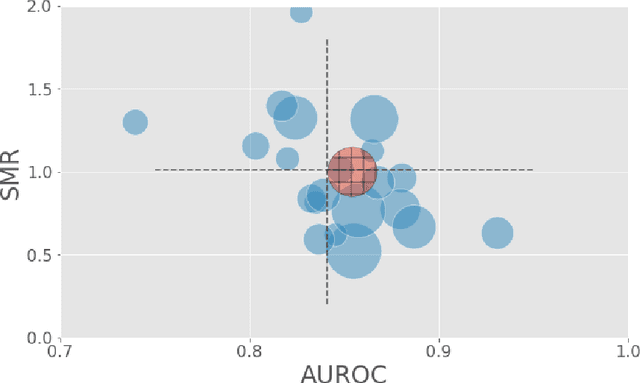
Abstract:A large volume of research has considered the creation of predictive models for clinical data; however, much existing literature reports results using only a single source of data. In this work, we evaluate the performance of models trained on the publicly-available eICU Collaborative Research Database. We show that cross-validation using many distinct centers provides a reasonable estimate of model performance in new centers. We further show that a single model trained across centers transfers well to distinct hospitals, even compared to a model retrained using hospital-specific data. Our results motivate the use of multi-center datasets for model development and highlight the need for data sharing among hospitals to maximize model performance.
False arrhythmia alarm reduction in the intensive care unit
Sep 11, 2017



Abstract:Research has shown that false alarms constitute more than 80% of the alarms triggered in the intensive care unit (ICU). The high false arrhythmia alarm rate has severe implications such as disruption of patient care, caregiver alarm fatigue, and desensitization from clinical staff to real life-threatening alarms. A method to reduce the false alarm rate would therefore greatly benefit patients as well as nurses in their ability to provide care. We here develop and describe a robust false arrhythmia alarm reduction system for use in the ICU. Building off of work previously described in the literature, we make use of signal processing and machine learning techniques to identify true and false alarms for five arrhythmia types. This baseline algorithm alone is able to perform remarkably well, with a sensitivity of 0.908, a specificity of 0.838, and a PhysioNet/CinC challenge score of 0.756. We additionally explore dynamic time warping techniques on both the entire alarm signal as well as on a beat-by-beat basis in an effort to improve performance of ventricular tachycardia, which has in the literature been one of the hardest arrhythmias to classify. Such an algorithm with strong performance and efficiency could potentially be translated for use in the ICU to promote overall patient care and recovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge