Steven Horng
Bidirectional Captioning for Clinically Accurate and Interpretable Models
Oct 30, 2023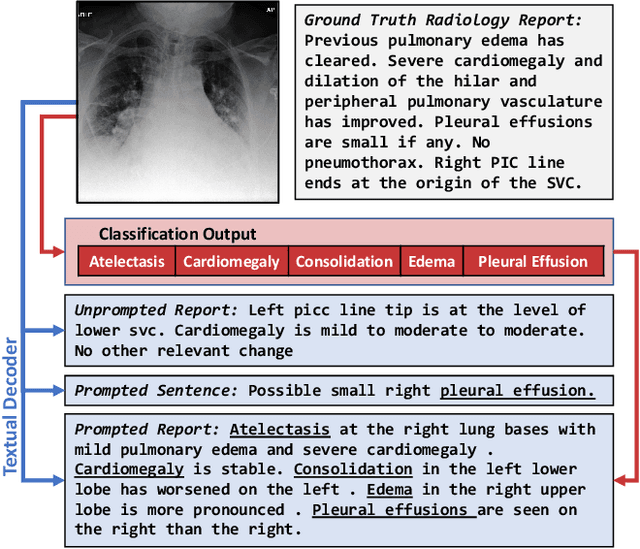
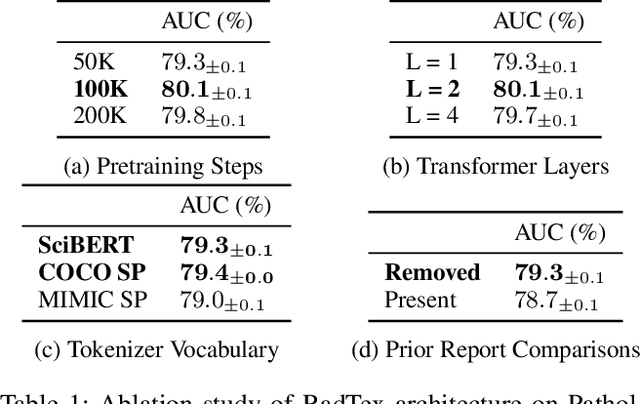
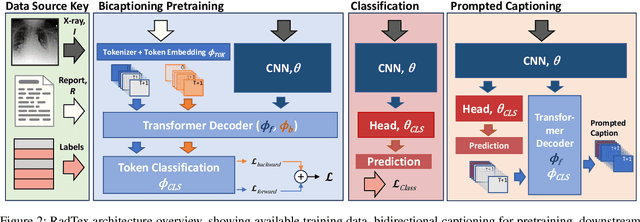
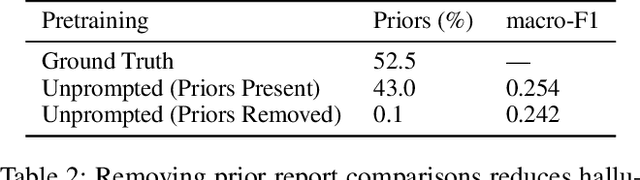
Abstract:Vision-language pretraining has been shown to produce high-quality visual encoders which transfer efficiently to downstream computer vision tasks. While generative language models have gained widespread attention, image captioning has thus far been mostly overlooked as a form of cross-modal pretraining in favor of contrastive learning, especially in medical image analysis. In this paper, we experiment with bidirectional captioning of radiology reports as a form of pretraining and compare the quality and utility of learned embeddings with those from contrastive pretraining methods. We optimize a CNN encoder, transformer decoder architecture named RadTex for the radiology domain. Results show that not only does captioning pretraining yield visual encoders that are competitive with contrastive pretraining (CheXpert competition multi-label AUC of 89.4%), but also that our transformer decoder is capable of generating clinically relevant reports (captioning macro-F1 score of 0.349 using CheXpert labeler) and responding to prompts with targeted, interactive outputs.
Conceptualizing Machine Learning for Dynamic Information Retrieval of Electronic Health Record Notes
Aug 09, 2023Abstract:The large amount of time clinicians spend sifting through patient notes and documenting in electronic health records (EHRs) is a leading cause of clinician burnout. By proactively and dynamically retrieving relevant notes during the documentation process, we can reduce the effort required to find relevant patient history. In this work, we conceptualize the use of EHR audit logs for machine learning as a source of supervision of note relevance in a specific clinical context, at a particular point in time. Our evaluation focuses on the dynamic retrieval in the emergency department, a high acuity setting with unique patterns of information retrieval and note writing. We show that our methods can achieve an AUC of 0.963 for predicting which notes will be read in an individual note writing session. We additionally conduct a user study with several clinicians and find that our framework can help clinicians retrieve relevant information more efficiently. Demonstrating that our framework and methods can perform well in this demanding setting is a promising proof of concept that they will translate to other clinical settings and data modalities (e.g., labs, medications, imaging).
Sample-Specific Debiasing for Better Image-Text Models
Apr 25, 2023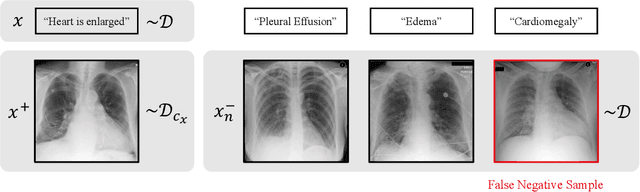
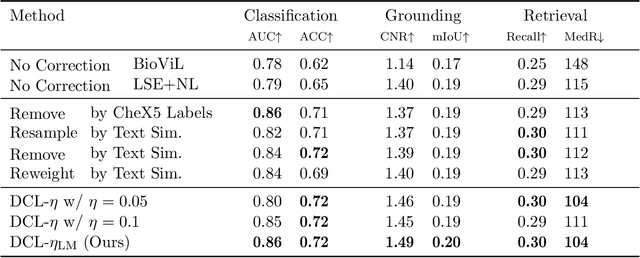
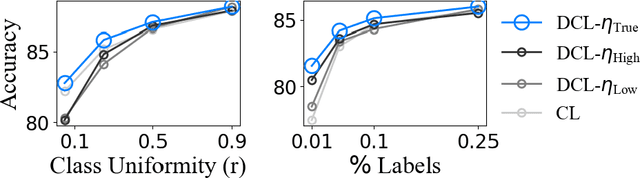
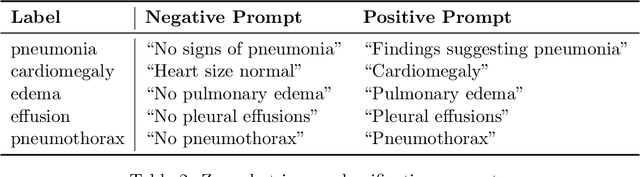
Abstract:Self-supervised representation learning on image-text data facilitates crucial medical applications, such as image classification, visual grounding, and cross-modal retrieval. One common approach involves contrasting semantically similar (positive) and dissimilar (negative) pairs of data points. Drawing negative samples uniformly from the training data set introduces false negatives, i.e., samples that are treated as dissimilar but belong to the same class. In healthcare data, the underlying class distribution is nonuniform, implying that false negatives occur at a highly variable rate. To improve the quality of learned representations, we develop a novel approach that corrects for false negatives. Our method can be viewed as a variant of debiased constrastive learning that uses estimated sample-specific class probabilities. We provide theoretical analysis of the objective function and demonstrate the proposed approach on both image and paired image-text data sets. Our experiments demonstrate empirical advantages of sample-specific debiasing.
Using Multiple Instance Learning to Build Multimodal Representations
Dec 11, 2022Abstract:Image-text multimodal representation learning aligns data across modalities and enables important medical applications, e.g., image classification, visual grounding, and cross-modal retrieval. In this work, we establish a connection between multimodal representation learning and multiple instance learning. Based on this connection, we propose a generic framework for constructing permutation-invariant score functions with many existing multimodal representation learning approaches as special cases. Furthermore, we use the framework to derive a novel contrastive learning approach and demonstrate that our method achieves state-of-the-art results on a number of downstream tasks.
RadTex: Learning Efficient Radiograph Representations from Text Reports
Aug 05, 2022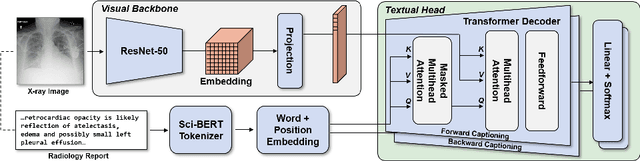
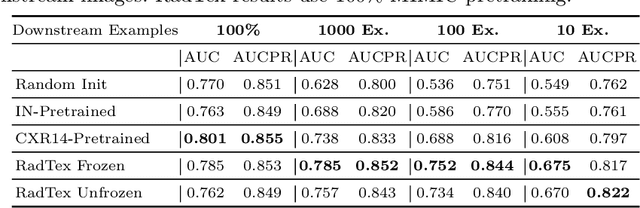
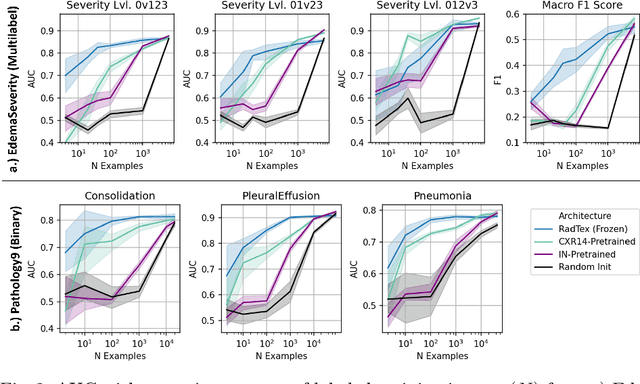
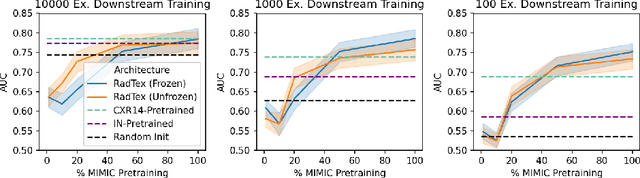
Abstract:Automated analysis of chest radiography using deep learning has tremendous potential to enhance the clinical diagnosis of diseases in patients. However, deep learning models typically require large amounts of annotated data to achieve high performance -- often an obstacle to medical domain adaptation. In this paper, we build a data-efficient learning framework that utilizes radiology reports to improve medical image classification performance with limited labeled data (fewer than 1000 examples). Specifically, we examine image-captioning pretraining to learn high-quality medical image representations that train on fewer examples. Following joint pretraining of a convolutional encoder and transformer decoder, we transfer the learned encoder to various classification tasks. Averaged over 9 pathologies, we find that our model achieves higher classification performance than ImageNet-supervised and in-domain supervised pretraining when labeled training data is limited.
Image Classification with Consistent Supporting Evidence
Nov 13, 2021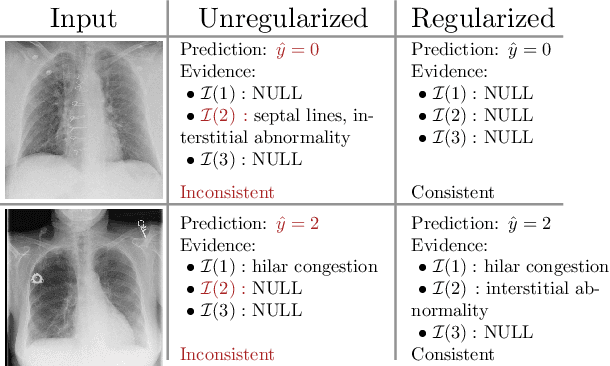
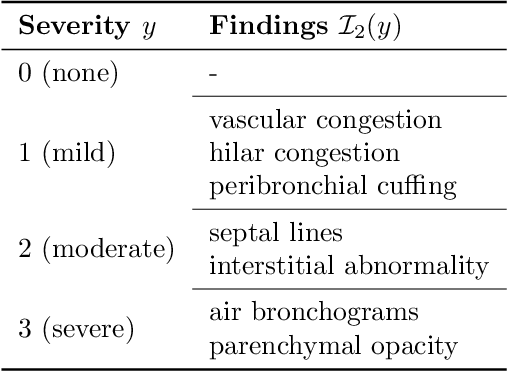
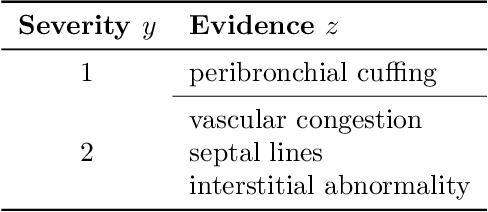
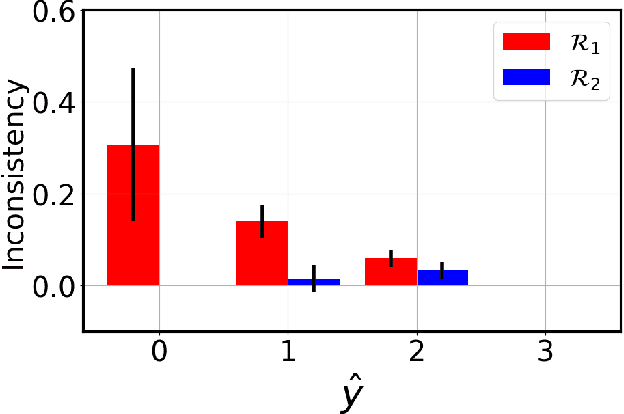
Abstract:Adoption of machine learning models in healthcare requires end users' trust in the system. Models that provide additional supportive evidence for their predictions promise to facilitate adoption. We define consistent evidence to be both compatible and sufficient with respect to model predictions. We propose measures of model inconsistency and regularizers that promote more consistent evidence. We demonstrate our ideas in the context of edema severity grading from chest radiographs. We demonstrate empirically that consistent models provide competitive performance while supporting interpretation.
Multimodal Representation Learning via Maximization of Local Mutual Information
Mar 08, 2021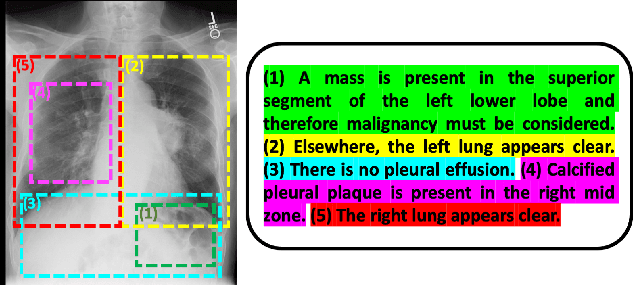
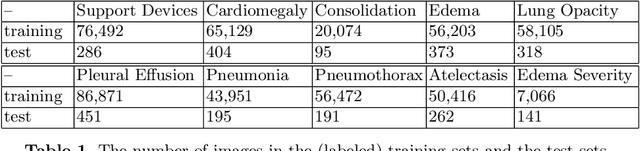
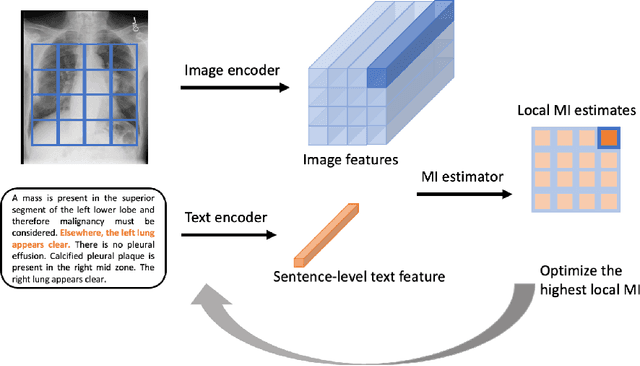
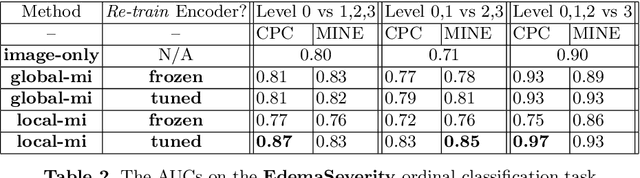
Abstract:We propose and demonstrate a representation learning approach by maximizing the mutual information between local features of images and text. The goal of this approach is to learn useful image representations by taking advantage of the rich information contained in the free text that describes the findings in the image. Our method learns image and text encoders by encouraging the resulting representations to exhibit high local mutual information. We make use of recent advances in mutual information estimation with neural network discriminators. We argue that, typically, the sum of local mutual information is a lower bound on the global mutual information. Our experimental results in the downstream image classification tasks demonstrate the advantages of using local features for image-text representation learning.
Joint Modeling of Chest Radiographs and Radiology Reports for Pulmonary Edema Assessment
Aug 22, 2020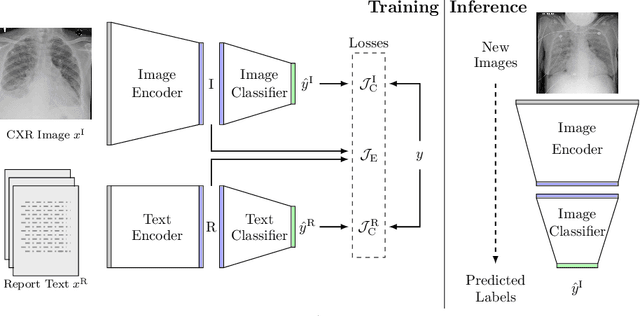
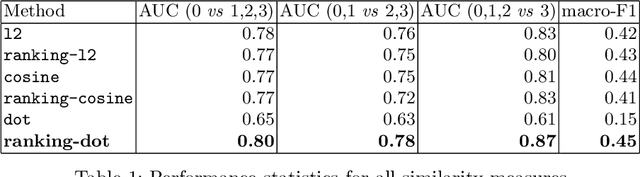

Abstract:We propose and demonstrate a novel machine learning algorithm that assesses pulmonary edema severity from chest radiographs. While large publicly available datasets of chest radiographs and free-text radiology reports exist, only limited numerical edema severity labels can be extracted from radiology reports. This is a significant challenge in learning such models for image classification. To take advantage of the rich information present in the radiology reports, we develop a neural network model that is trained on both images and free-text to assess pulmonary edema severity from chest radiographs at inference time. Our experimental results suggest that the joint image-text representation learning improves the performance of pulmonary edema assessment compared to a supervised model trained on images only. We also show the use of the text for explaining the image classification by the joint model. To the best of our knowledge, our approach is the first to leverage free-text radiology reports for improving the image model performance in this application. Our code is available at https://github.com/RayRuizhiLiao/joint_chestxray.
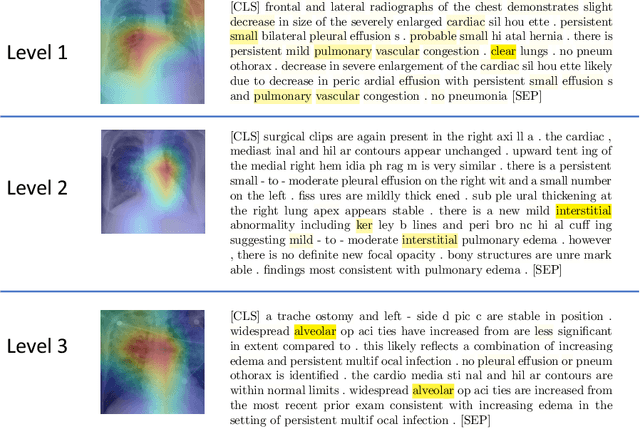
Deep Learning to Quantify Pulmonary Edema in Chest Radiographs
Aug 13, 2020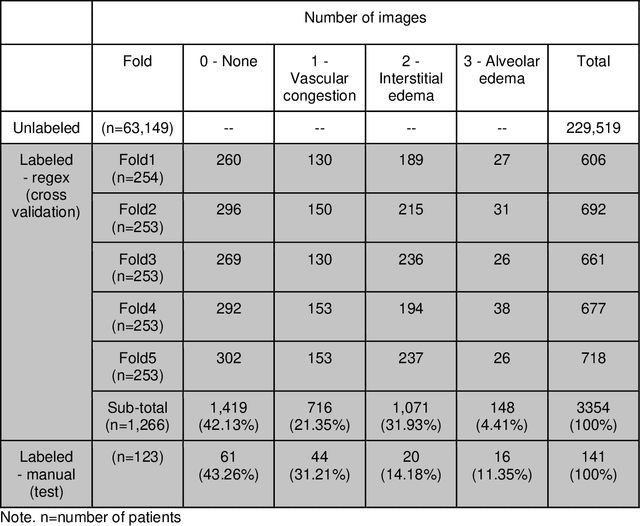
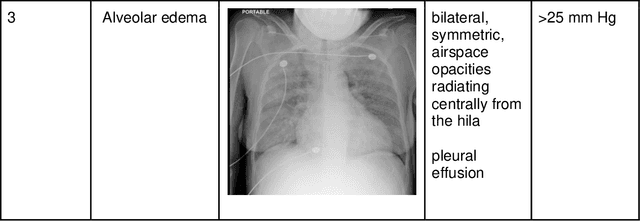
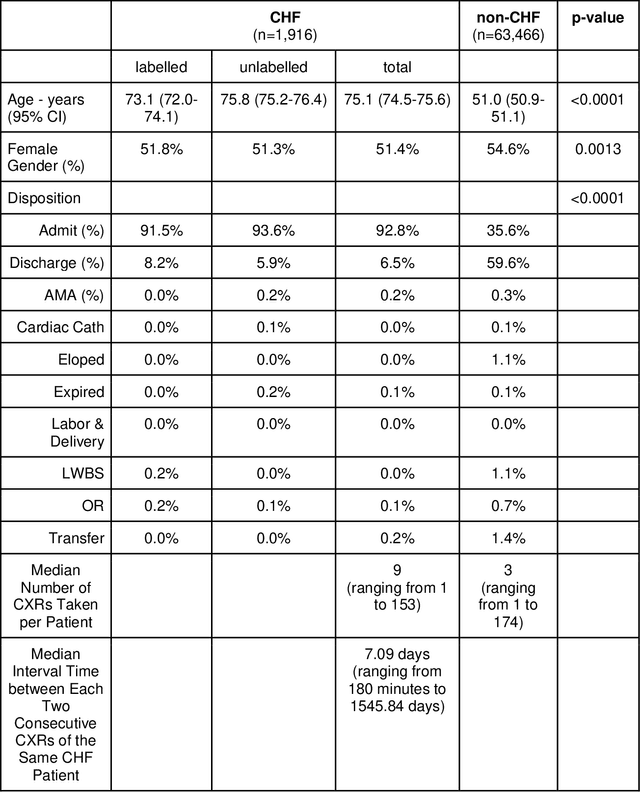
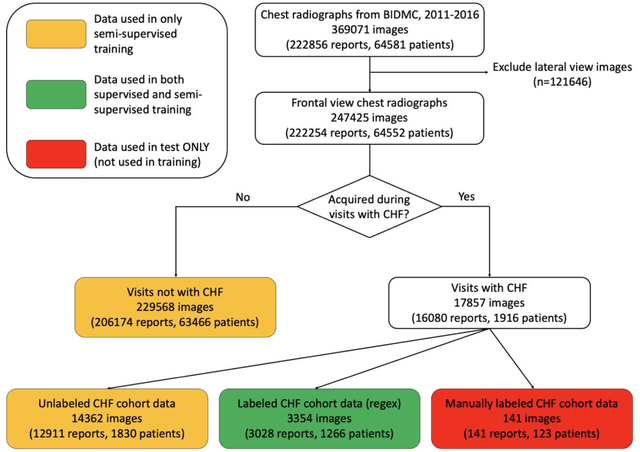
Abstract:Background: Clinical management decisions for acutely decompensated CHF patients are often based on grades of pulmonary edema severity, rather than its mere absence or presence. The grading of pulmonary edema on chest radiographs is based on well-known radiologic findings. Purpose: We develop a clinical machine learning task to grade pulmonary edema severity and release both the underlying data and code to serve as a benchmark for future algorithmic developments in machine vision. Materials and Methods: We collected 369,071 chest radiographs and their associated radiology reports from 64,581 patients from the MIMIC-CXR chest radiograph dataset. We extracted pulmonary edema severity labels from the associated radiology reports as 4 ordinal levels: no edema (0), vascular congestion (1), interstitial edema (2), and alveolar edema (3). We developed machine learning models using two standard approaches: 1) a semi-supervised model using a variational autoencoder and 2) a pre-trained supervised learning model using a dense neural network. Results: We measured the area under the receiver operating characteristic curve (AUROC) from the semi-supervised model and the pre-trained model. AUROC for differentiating alveolar edema from no edema was 0.99 and 0.87 (semi-supervised and pre-trained models). Performance of the algorithm was inversely related to the difficulty in categorizing milder states of pulmonary edema: 2 vs 0 (0.88, 0.81), 1 vs 0 (0.79, 0.66), 3 vs 1 (0.93, 0.82), 2 vs 1 (0.69, 0.73), 3 vs 2 (0.88, 0.63). Conclusion: Accurate grading of pulmonary edema on chest radiographs is a clinically important task. Application of state-of-the-art machine learning techniques can produce a novel quantitative imaging biomarker from one of the oldest and most widely available imaging modalities.
Fast, Structured Clinical Documentation via Contextual Autocomplete
Jul 29, 2020



Abstract:We present a system that uses a learned autocompletion mechanism to facilitate rapid creation of semi-structured clinical documentation. We dynamically suggest relevant clinical concepts as a doctor drafts a note by leveraging features from both unstructured and structured medical data. By constraining our architecture to shallow neural networks, we are able to make these suggestions in real time. Furthermore, as our algorithm is used to write a note, we can automatically annotate the documentation with clean labels of clinical concepts drawn from medical vocabularies, making notes more structured and readable for physicians, patients, and future algorithms. To our knowledge, this system is the only machine learning-based documentation utility for clinical notes deployed in a live hospital setting, and it reduces keystroke burden of clinical concepts by 67% in real environments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge