Peter Szolovits
N3C Natural Language Processing
Right, No Matter Why: AI Fact-checking and AI Authority in Health-related Inquiry Settings
Oct 22, 2023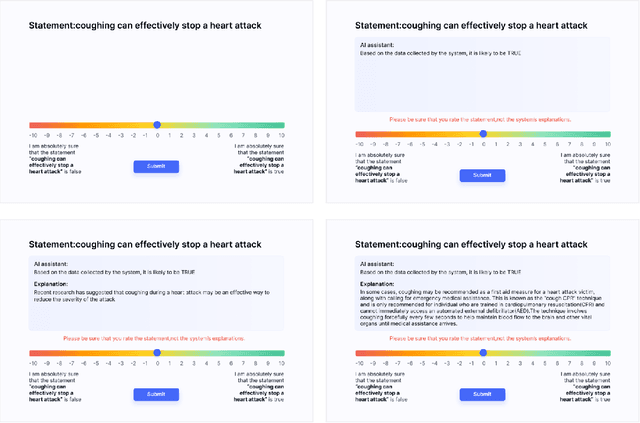
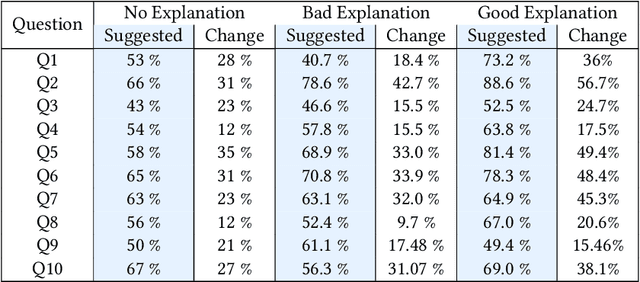
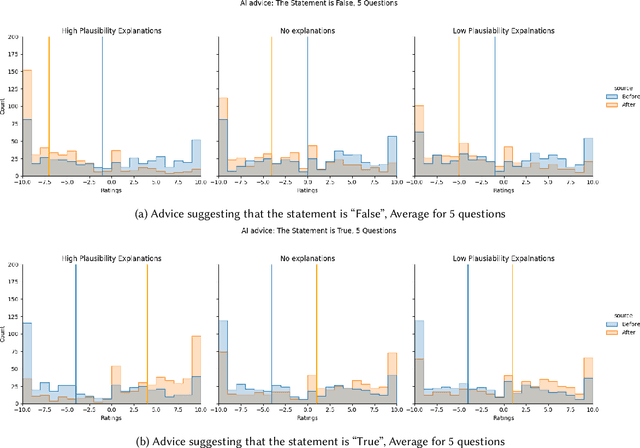

Abstract:Previous research on expert advice-taking shows that humans exhibit two contradictory behaviors: on the one hand, people tend to overvalue their own opinions undervaluing the expert opinion, and on the other, people often defer to other people's advice even if the advice itself is rather obviously wrong. In our study, we conduct an exploratory evaluation of users' AI-advice accepting behavior when evaluating the truthfulness of a health-related statement in different "advice quality" settings. We find that even feedback that is confined to just stating that "the AI thinks that the statement is false/true" results in more than half of people moving their statement veracity assessment towards the AI suggestion. The different types of advice given influence the acceptance rates, but the sheer effect of getting a suggestion is often bigger than the suggestion-type effect.
Do We Still Need Clinical Language Models?
Feb 16, 2023



Abstract:Although recent advances in scaling large language models (LLMs) have resulted in improvements on many NLP tasks, it remains unclear whether these models trained primarily with general web text are the right tool in highly specialized, safety critical domains such as clinical text. Recent results have suggested that LLMs encode a surprising amount of medical knowledge. This raises an important question regarding the utility of smaller domain-specific language models. With the success of general-domain LLMs, is there still a need for specialized clinical models? To investigate this question, we conduct an extensive empirical analysis of 12 language models, ranging from 220M to 175B parameters, measuring their performance on 3 different clinical tasks that test their ability to parse and reason over electronic health records. As part of our experiments, we train T5-Base and T5-Large models from scratch on clinical notes from MIMIC III and IV to directly investigate the efficiency of clinical tokens. We show that relatively small specialized clinical models substantially outperform all in-context learning approaches, even when finetuned on limited annotated data. Further, we find that pretraining on clinical tokens allows for smaller, more parameter-efficient models that either match or outperform much larger language models trained on general text. We release the code and the models used under the PhysioNet Credentialed Health Data license and data use agreement.
Using Machine Learning to Develop Smart Reflex Testing Protocols
Feb 01, 2023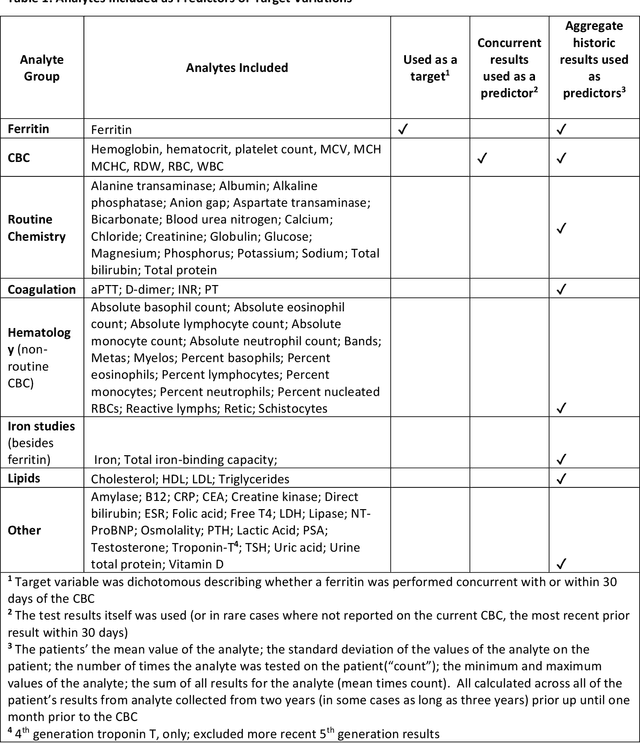
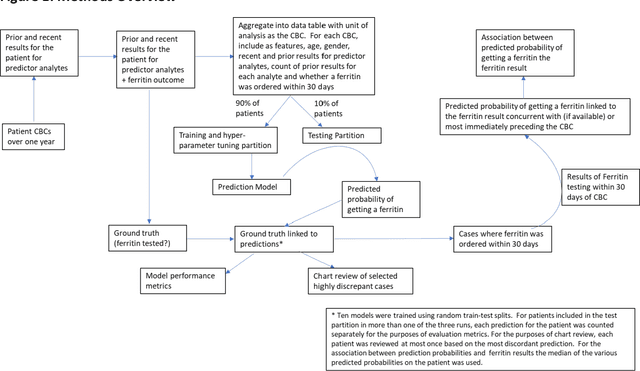
Abstract:Objective: Reflex testing protocols allow clinical laboratories to perform second line diagnostic tests on existing specimens based on the results of initially ordered tests. Reflex testing can support optimal clinical laboratory test ordering and diagnosis. In current clinical practice, reflex testing typically relies on simple "if-then" rules; however, this limits their scope since most test ordering decisions involve more complexity than a simple rule will allow. Here, using the analyte ferritin as an example, we propose an alternative machine learning-based approach to "smart" reflex testing with a wider scope and greater impact than traditional rule-based approaches. Methods: Using patient data, we developed a machine learning model to predict whether a patient getting CBC testing will also have ferritin testing ordered, consider applications of this model to "smart" reflex testing, and evaluate the model by comparing its performance to possible rule-based approaches. Results: Our underlying machine learning models performed moderately well in predicting ferritin test ordering and demonstrated greater suitability to reflex testing than rule-based approaches. Using chart review, we demonstrate that our model may improve ferritin test ordering. Finally, as a secondary goal, we demonstrate that ferritin test results are missing not at random (MNAR), a finding with implications for unbiased imputation of missing test results. Conclusions: Machine learning may provide a foundation for new types of reflex testing with enhanced benefits for clinical diagnosis and laboratory utilization management.
Learning to Ask Like a Physician
Jun 06, 2022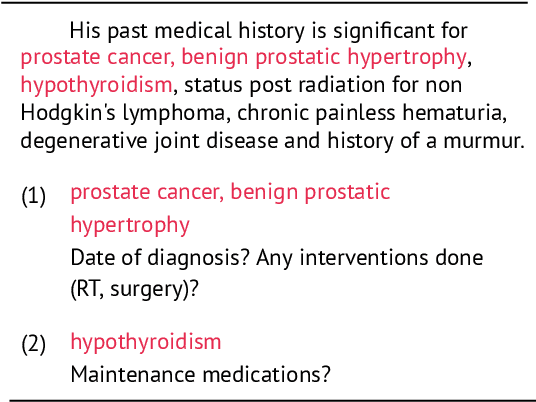
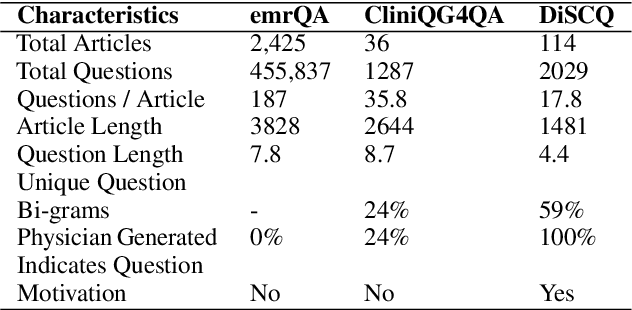
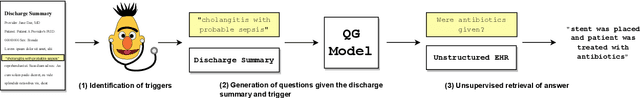
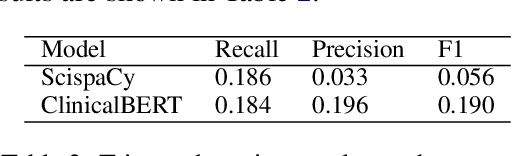
Abstract:Existing question answering (QA) datasets derived from electronic health records (EHR) are artificially generated and consequently fail to capture realistic physician information needs. We present Discharge Summary Clinical Questions (DiSCQ), a newly curated question dataset composed of 2,000+ questions paired with the snippets of text (triggers) that prompted each question. The questions are generated by medical experts from 100+ MIMIC-III discharge summaries. We analyze this dataset to characterize the types of information sought by medical experts. We also train baseline models for trigger detection and question generation (QG), paired with unsupervised answer retrieval over EHRs. Our baseline model is able to generate high quality questions in over 62% of cases when prompted with human selected triggers. We release this dataset (and all code to reproduce baseline model results) to facilitate further research into realistic clinical QA and QG: https://github.com/elehman16/discq.
Explainable Deep Learning in Healthcare: A Methodological Survey from an Attribution View
Dec 05, 2021
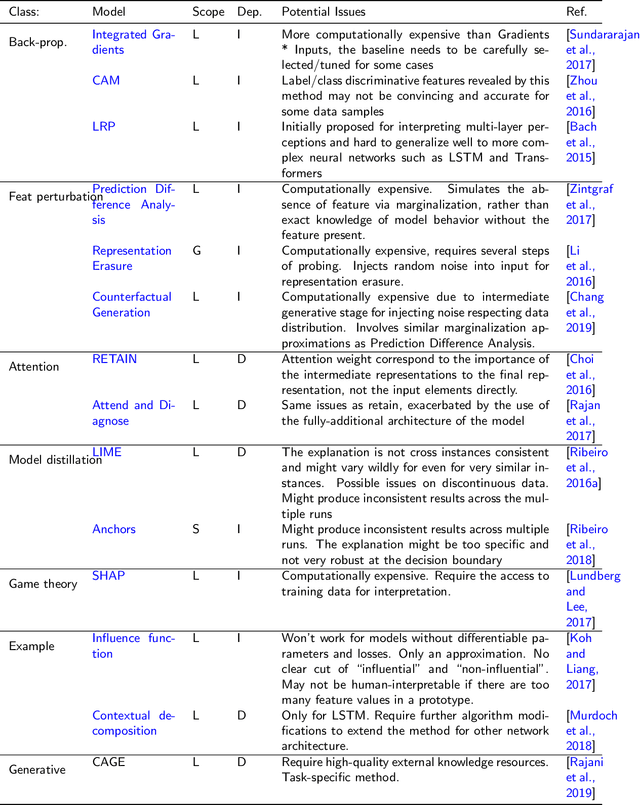
Abstract:The increasing availability of large collections of electronic health record (EHR) data and unprecedented technical advances in deep learning (DL) have sparked a surge of research interest in developing DL based clinical decision support systems for diagnosis, prognosis, and treatment. Despite the recognition of the value of deep learning in healthcare, impediments to further adoption in real healthcare settings remain due to the black-box nature of DL. Therefore, there is an emerging need for interpretable DL, which allows end users to evaluate the model decision making to know whether to accept or reject predictions and recommendations before an action is taken. In this review, we focus on the interpretability of the DL models in healthcare. We start by introducing the methods for interpretability in depth and comprehensively as a methodological reference for future researchers or clinical practitioners in this field. Besides the methods' details, we also include a discussion of advantages and disadvantages of these methods and which scenarios each of them is suitable for, so that interested readers can know how to compare and choose among them for use. Moreover, we discuss how these methods, originally developed for solving general-domain problems, have been adapted and applied to healthcare problems and how they can help physicians better understand these data-driven technologies. Overall, we hope this survey can help researchers and practitioners in both artificial intelligence (AI) and clinical fields understand what methods we have for enhancing the interpretability of their DL models and choose the optimal one accordingly.
An Open Natural Language Processing Development Framework for EHR-based Clinical Research: A case demonstration using the National COVID Cohort Collaborative (N3C)
Oct 20, 2021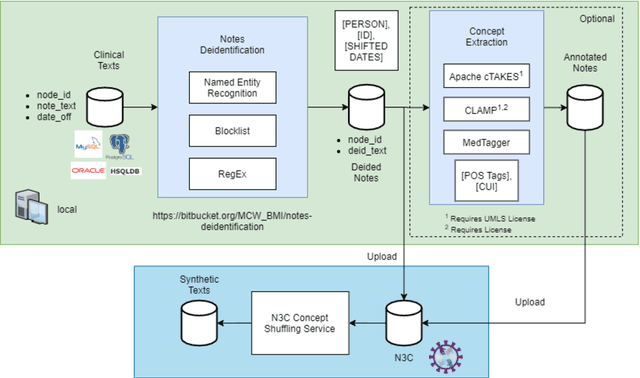
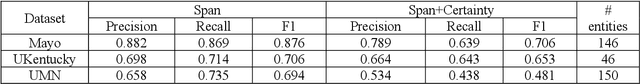
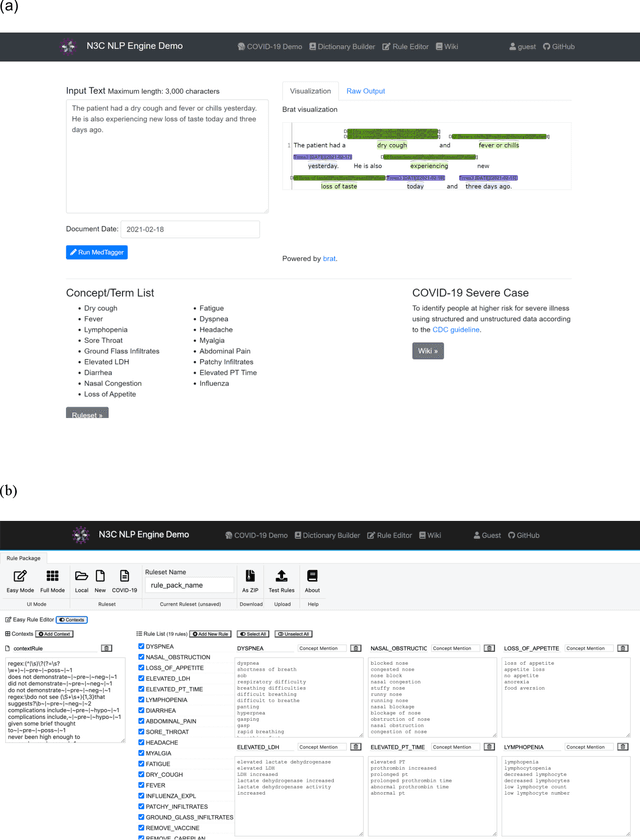
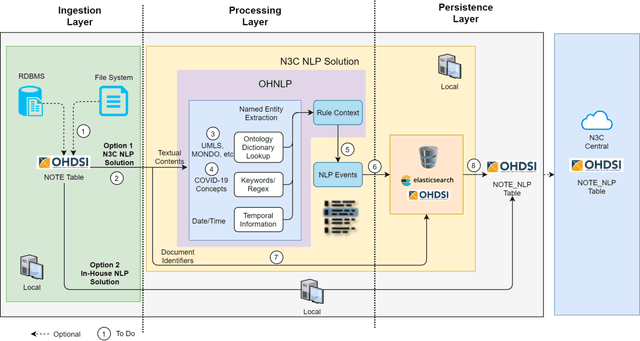
Abstract:While we pay attention to the latest advances in clinical natural language processing (NLP), we can notice some resistance in the clinical and translational research community to adopt NLP models due to limited transparency, Interpretability and usability. Built upon our previous work, in this study, we proposed an open natural language processing development framework and evaluated it through the implementation of NLP algorithms for the National COVID Cohort Collaborative (N3C). Based on the interests in information extraction from COVID-19 related clinical notes, our work includes 1) an open data annotation process using COVID-19 signs and symptoms as the use case, 2) a community-driven ruleset composing platform, and 3) a synthetic text data generation workflow to generate texts for information extraction tasks without involving human subjects. The generated corpora derived out of the texts from multiple intuitions and gold standard annotation are tested on a single institution's rule set has the performances in F1 score of 0.876, 0.706 and 0.694, respectively. The study as a consortium effort of the N3C NLP subgroup demonstrates the feasibility of creating a federated NLP algorithm development and benchmarking platform to enhance multi-institution clinical NLP study.
Rethinking Relational Encoding in Language Model: Pre-Training for General Sequences
Mar 18, 2021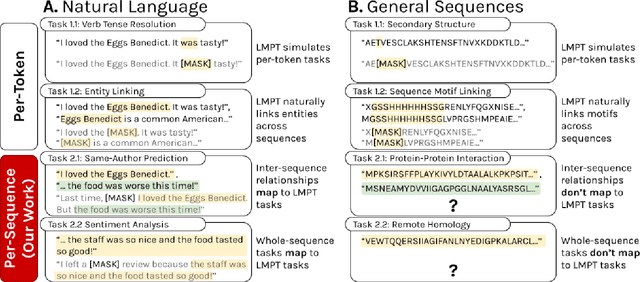
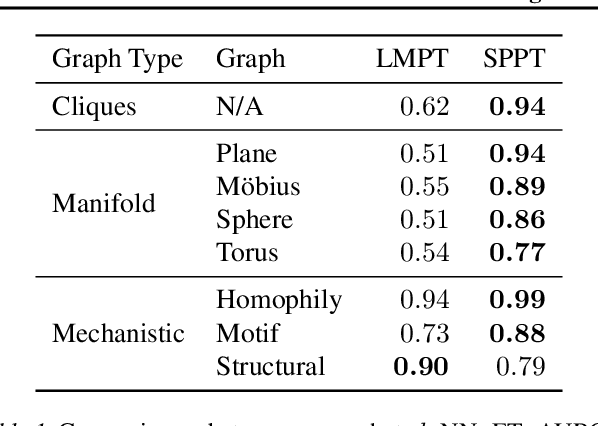
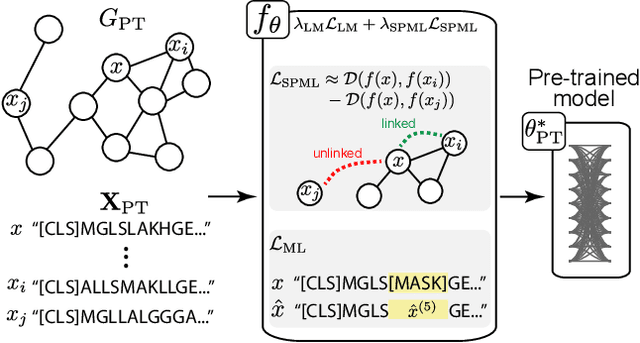
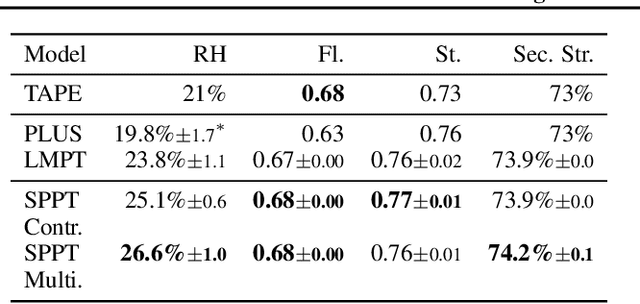
Abstract:Language model pre-training (LMPT) has achieved remarkable results in natural language understanding. However, LMPT is much less successful in non-natural language domains like protein sequences, revealing a crucial discrepancy between the various sequential domains. Here, we posit that while LMPT can effectively model per-token relations, it fails at modeling per-sequence relations in non-natural language domains. To this end, we develop a framework that couples LMPT with deep structure-preserving metric learning to produce richer embeddings than can be obtained from LMPT alone. We examine new and existing pre-training models in this framework and theoretically analyze the framework overall. We also design experiments on a variety of synthetic datasets and new graph-augmented datasets of proteins and scientific abstracts. Our approach offers notable performance improvements on downstream tasks, including prediction of protein remote homology and classification of citation intent.
Adversarial Contrastive Pre-training for Protein Sequences
Jan 31, 2021


Abstract:Recent developments in Natural Language Processing (NLP) demonstrate that large-scale, self-supervised pre-training can be extremely beneficial for downstream tasks. These ideas have been adapted to other domains, including the analysis of the amino acid sequences of proteins. However, to date most attempts on protein sequences rely on direct masked language model style pre-training. In this work, we design a new, adversarial pre-training method for proteins, extending and specializing similar advances in NLP. We show compelling results in comparison to traditional MLM pre-training, though further development is needed to ensure the gains are worth the significant computational cost.
What Disease does this Patient Have? A Large-scale Open Domain Question Answering Dataset from Medical Exams
Sep 28, 2020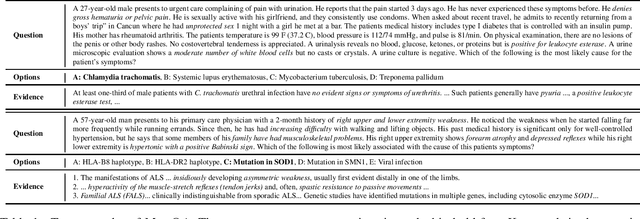
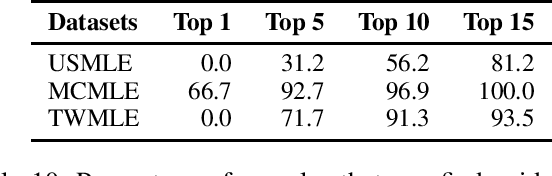

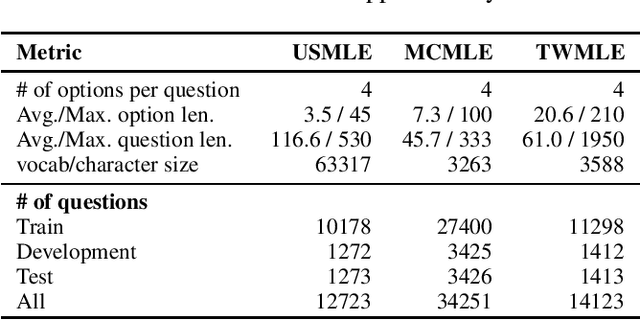
Abstract:Open domain question answering (OpenQA) tasks have been recently attracting more and more attention from the natural language processing (NLP) community. In this work, we present the first free-form multiple-choice OpenQA dataset for solving medical problems, MedQA, collected from the professional medical board exams. It covers three languages: English, simplified Chinese, and traditional Chinese, and contains 12,723, 34,251, and 14,123 questions for the three languages, respectively. We implement both rule-based and popular neural methods by sequentially combining a document retriever and a machine comprehension model. Through experiments, we find that even the current best method can only achieve 36.7\%, 42.0\%, and 70.1\% of test accuracy on the English, traditional Chinese, and simplified Chinese questions, respectively. We expect MedQA to present great challenges to existing OpenQA systems and hope that it can serve as a platform to promote much stronger OpenQA models from the NLP community in the future.
Joint Modeling of Chest Radiographs and Radiology Reports for Pulmonary Edema Assessment
Aug 22, 2020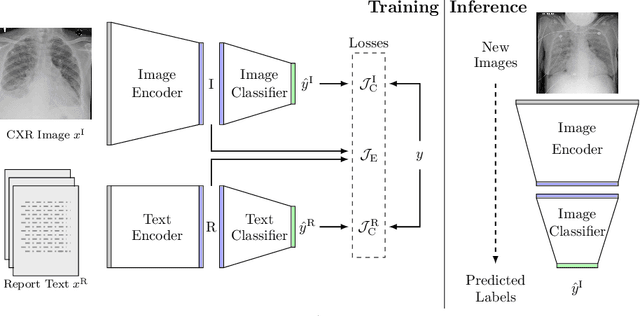
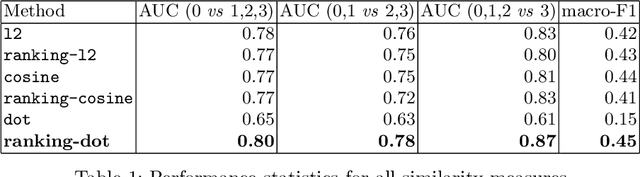

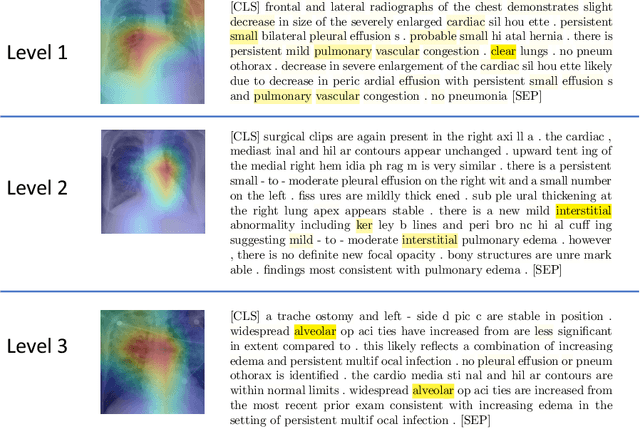
Abstract:We propose and demonstrate a novel machine learning algorithm that assesses pulmonary edema severity from chest radiographs. While large publicly available datasets of chest radiographs and free-text radiology reports exist, only limited numerical edema severity labels can be extracted from radiology reports. This is a significant challenge in learning such models for image classification. To take advantage of the rich information present in the radiology reports, we develop a neural network model that is trained on both images and free-text to assess pulmonary edema severity from chest radiographs at inference time. Our experimental results suggest that the joint image-text representation learning improves the performance of pulmonary edema assessment compared to a supervised model trained on images only. We also show the use of the text for explaining the image classification by the joint model. To the best of our knowledge, our approach is the first to leverage free-text radiology reports for improving the image model performance in this application. Our code is available at https://github.com/RayRuizhiLiao/joint_chestxray.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge