Marinka Zitnik
Department of Biomedical Informatics, Harvard Medical School, Boston, MA, USA, Kempner Institute for the Study of Natural and Artificial Intelligence, Allston, MA, USA, Broad Institute of MIT and Harvard, Cambridge, MA, USA, Harvard Data Science Initiative, Cambridge, MA, USA
Autonomous Knowledge Graph Exploration with Adaptive Breadth-Depth Retrieval
Jan 20, 2026Abstract:Retrieving evidence for language model queries from knowledge graphs requires balancing broad search across the graph with multi-hop traversal to follow relational links. Similarity-based retrievers provide coverage but remain shallow, whereas traversal-based methods rely on selecting seed nodes to start exploration, which can fail when queries span multiple entities and relations. We introduce ARK: Adaptive Retriever of Knowledge, an agentic KG retriever that gives a language model control over this breadth-depth tradeoff using a two-operation toolset: global lexical search over node descriptors and one-hop neighborhood exploration that composes into multi-hop traversal. ARK alternates between breadth-oriented discovery and depth-oriented expansion without depending on a fragile seed selection, a pre-set hop depth, or requiring retrieval training. ARK adapts tool use to queries, using global search for language-heavy queries and neighborhood exploration for relation-heavy queries. On STaRK, ARK reaches 59.1% average Hit@1 and 67.4 average MRR, improving average Hit@1 by up to 31.4% and average MRR by up to 28.0% over retrieval-based and agentic training-free methods. Finally, we distill ARK's tool-use trajectories from a large teacher into an 8B model via label-free imitation, improving Hit@1 by +7.0, +26.6, and +13.5 absolute points over the base 8B model on AMAZON, MAG, and PRIME datasets, respectively, while retaining up to 98.5% of the teacher's Hit@1 rate.
Beyond Affinity: A Benchmark of 1D, 2D, and 3D Methods Reveals Critical Trade-offs in Structure-Based Drug Design
Jan 13, 2026Abstract:Currently, the field of structure-based drug design is dominated by three main types of algorithms: search-based algorithms, deep generative models, and reinforcement learning. While existing works have typically focused on comparing models within a single algorithmic category, cross-algorithm comparisons remain scarce. In this paper, to fill the gap, we establish a benchmark to evaluate the performance of fifteen models across these different algorithmic foundations by assessing the pharmaceutical properties of the generated molecules and their docking affinities and poses with specified target proteins. We highlight the unique advantages of each algorithmic approach and offer recommendations for the design of future SBDD models. We emphasize that 1D/2D ligand-centric drug design methods can be used in SBDD by treating the docking function as a black-box oracle, which is typically neglected. Our evaluation reveals distinct patterns across model categories. 3D structure-based models excel in binding affinities but show inconsistencies in chemical validity and pose quality. 1D models demonstrate reliable performance in standard molecular metrics but rarely achieve optimal binding affinities. 2D models offer balanced performance, maintaining high chemical validity while achieving moderate binding scores. Through detailed analysis across multiple protein targets, we identify key improvement areas for each model category, providing insights for researchers to combine strengths of different approaches while addressing their limitations. All the code that are used for benchmarking is available in https://github.com/zkysfls/2025-sbdd-benchmark
Graph AI generates neurological hypotheses validated in molecular, organoid, and clinical systems
Dec 13, 2025Abstract:Neurological diseases are the leading global cause of disability, yet most lack disease-modifying treatments. We present PROTON, a heterogeneous graph transformer that generates testable hypotheses across molecular, organoid, and clinical systems. To evaluate PROTON, we apply it to Parkinson's disease (PD), bipolar disorder (BD), and Alzheimer's disease (AD). In PD, PROTON linked genetic risk loci to genes essential for dopaminergic neuron survival and predicted pesticides toxic to patient-derived neurons, including the insecticide endosulfan, which ranked within the top 1.29% of predictions. In silico screens performed by PROTON reproduced six genome-wide $α$-synuclein experiments, including a split-ubiquitin yeast two-hybrid system (normalized enrichment score [NES] = 2.30, FDR-adjusted $p < 1 \times 10^{-4}$), an ascorbate peroxidase proximity labeling assay (NES = 2.16, FDR $< 1 \times 10^{-4}$), and a high-depth targeted exome sequencing study in 496 synucleinopathy patients (NES = 2.13, FDR $< 1 \times 10^{-4}$). In BD, PROTON predicted calcitriol as a candidate drug that reversed proteomic alterations observed in cortical organoids derived from BD patients. In AD, we evaluated PROTON predictions in health records from $n = 610,524$ patients at Mass General Brigham, confirming that five PROTON-predicted drugs were associated with reduced seven-year dementia risk (minimum hazard ratio = 0.63, 95% CI: 0.53-0.75, $p < 1 \times 10^{-7}$). PROTON generated neurological hypotheses that were evaluated across molecular, organoid, and clinical systems, defining a path for AI-driven discovery in neurological disease.
Protein Structure Tokenization via Geometric Byte Pair Encoding
Nov 13, 2025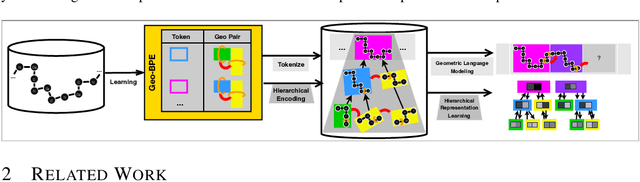
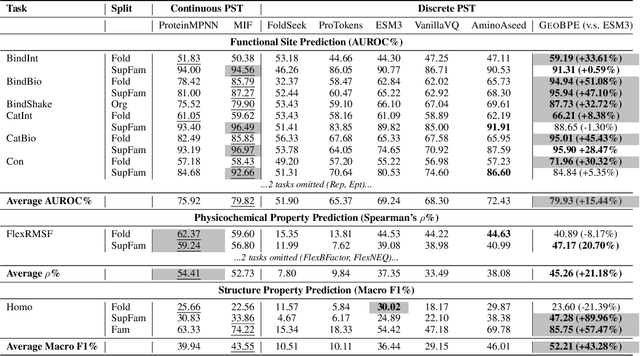
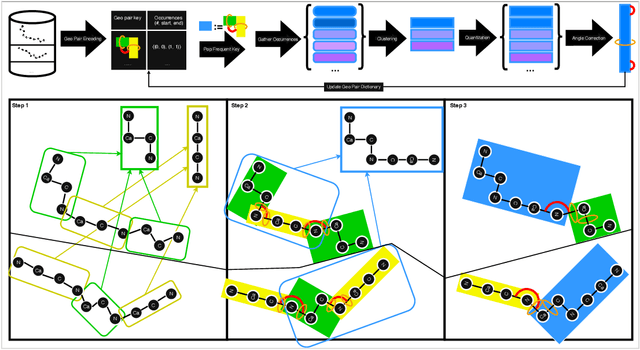
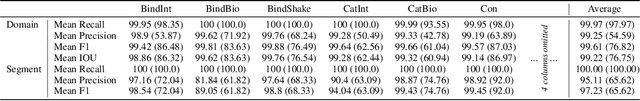
Abstract:Protein structure is central to biological function, and enabling multimodal protein models requires joint reasoning over sequence, structure, and function. A key barrier is the lack of principled protein structure tokenizers (PSTs): existing approaches fix token size or rely on continuous vector codebooks, limiting interpretability, multi-scale control, and transfer across architectures. We introduce GeoBPE, a geometry-grounded PST that transforms continuous, noisy, multi-scale backbone conformations into discrete ``sentences'' of geometry while enforcing global constraints. Analogous to byte-pair encoding, GeoBPE generates a hierarchical vocabulary of geometric primitives by iteratively (i) clustering Geo-Pair occurrences with k-medoids to yield a resolution-controllable vocabulary; (ii) quantizing each Geo-Pair to its closest medoid prototype; and (iii) reducing drift through differentiable inverse kinematics that optimizes boundary glue angles under an $\mathrm{SE}(3)$ end-frame loss. GeoBPE offers compression ($>$10x reduction in bits-per-residue at similar distortion rate), data efficiency ($>$10x less training data), and generalization (maintains test/train distortion ratio of $1.0-1.1$). It is architecture-agnostic: (a) its hierarchical vocabulary provides a strong inductive bias for coarsening residue-level embeddings from large PLMs into motif- and protein-level representations, consistently outperforming leading PSTs across $12$ tasks and $24$ test splits; (b) paired with a transformer, GeoBPE supports unconditional backbone generation via language modeling; and (c) tokens align with CATH functional families and support expert-interpretable case studies, offering functional meaning absent in prior PSTs. Code is available at https://github.com/shiningsunnyday/PT-BPE/.
One Patient, Many Contexts: Scaling Medical AI Through Contextual Intelligence
Jun 11, 2025

Abstract:Medical foundation models, including language models trained on clinical notes, vision-language models on medical images, and multimodal models on electronic health records, can summarize clinical notes, answer medical questions, and assist in decision-making. Adapting these models to new populations, specialties, or settings typically requires fine-tuning, careful prompting, or retrieval from knowledge bases. This can be impractical, and limits their ability to interpret unfamiliar inputs and adjust to clinical situations not represented during training. As a result, models are prone to contextual errors, where predictions appear reasonable but fail to account for critical patient-specific or contextual information. These errors stem from a fundamental limitation that current models struggle with: dynamically adjusting their behavior across evolving contexts of medical care. In this Perspective, we outline a vision for context-switching in medical AI: models that dynamically adapt their reasoning without retraining to new specialties, populations, workflows, and clinical roles. We envision context-switching AI to diagnose, manage, and treat a wide range of diseases across specialties and regions, and expand access to medical care.
Prompting Decision Transformers for Zero-Shot Reach-Avoid Policies
May 25, 2025Abstract:Offline goal-conditioned reinforcement learning methods have shown promise for reach-avoid tasks, where an agent must reach a target state while avoiding undesirable regions of the state space. Existing approaches typically encode avoid-region information into an augmented state space and cost function, which prevents flexible, dynamic specification of novel avoid-region information at evaluation time. They also rely heavily on well-designed reward and cost functions, limiting scalability to complex or poorly structured environments. We introduce RADT, a decision transformer model for offline, reward-free, goal-conditioned, avoid region-conditioned RL. RADT encodes goals and avoid regions directly as prompt tokens, allowing any number of avoid regions of arbitrary size to be specified at evaluation time. Using only suboptimal offline trajectories from a random policy, RADT learns reach-avoid behavior through a novel combination of goal and avoid-region hindsight relabeling. We benchmark RADT against 3 existing offline goal-conditioned RL models across 11 tasks, environments, and experimental settings. RADT generalizes in a zero-shot manner to out-of-distribution avoid region sizes and counts, outperforming baselines that require retraining. In one such zero-shot setting, RADT achieves 35.7% improvement in normalized cost over the best retrained baseline while maintaining high goal-reaching success. We apply RADT to cell reprogramming in biology, where it reduces visits to undesirable intermediate gene expression states during trajectories to desired target states, despite stochastic transitions and discrete, structured state dynamics.
Token Reduction Should Go Beyond Efficiency in Generative Models -- From Vision, Language to Multimodality
May 23, 2025Abstract:In Transformer architectures, tokens\textemdash discrete units derived from raw data\textemdash are formed by segmenting inputs into fixed-length chunks. Each token is then mapped to an embedding, enabling parallel attention computations while preserving the input's essential information. Due to the quadratic computational complexity of transformer self-attention mechanisms, token reduction has primarily been used as an efficiency strategy. This is especially true in single vision and language domains, where it helps balance computational costs, memory usage, and inference latency. Despite these advances, this paper argues that token reduction should transcend its traditional efficiency-oriented role in the era of large generative models. Instead, we position it as a fundamental principle in generative modeling, critically influencing both model architecture and broader applications. Specifically, we contend that across vision, language, and multimodal systems, token reduction can: (i) facilitate deeper multimodal integration and alignment, (ii) mitigate "overthinking" and hallucinations, (iii) maintain coherence over long inputs, and (iv) enhance training stability, etc. We reframe token reduction as more than an efficiency measure. By doing so, we outline promising future directions, including algorithm design, reinforcement learning-guided token reduction, token optimization for in-context learning, and broader ML and scientific domains. We highlight its potential to drive new model architectures and learning strategies that improve robustness, increase interpretability, and better align with the objectives of generative modeling.
PyTDC: A multimodal machine learning training, evaluation, and inference platform for biomedical foundation models
May 08, 2025
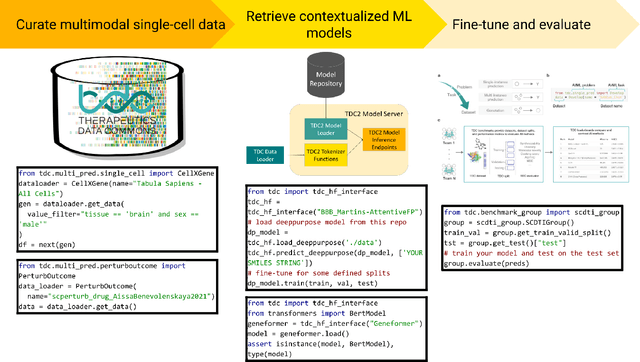


Abstract:Existing biomedical benchmarks do not provide end-to-end infrastructure for training, evaluation, and inference of models that integrate multimodal biological data and a broad range of machine learning tasks in therapeutics. We present PyTDC, an open-source machine-learning platform providing streamlined training, evaluation, and inference software for multimodal biological AI models. PyTDC unifies distributed, heterogeneous, continuously updated data sources and model weights and standardizes benchmarking and inference endpoints. This paper discusses the components of PyTDC's architecture and, to our knowledge, the first-of-its-kind case study on the introduced single-cell drug-target nomination ML task. We find state-of-the-art methods in graph representation learning and domain-specific methods from graph theory perform poorly on this task. Though we find a context-aware geometric deep learning method that outperforms the evaluated SoTA and domain-specific baseline methods, the model is unable to generalize to unseen cell types or incorporate additional modalities, highlighting PyTDC's capacity to facilitate an exciting avenue of research developing multimodal, context-aware, foundation models for open problems in biomedical AI.
TxAgent: An AI Agent for Therapeutic Reasoning Across a Universe of Tools
Mar 14, 2025Abstract:Precision therapeutics require multimodal adaptive models that generate personalized treatment recommendations. We introduce TxAgent, an AI agent that leverages multi-step reasoning and real-time biomedical knowledge retrieval across a toolbox of 211 tools to analyze drug interactions, contraindications, and patient-specific treatment strategies. TxAgent evaluates how drugs interact at molecular, pharmacokinetic, and clinical levels, identifies contraindications based on patient comorbidities and concurrent medications, and tailors treatment strategies to individual patient characteristics. It retrieves and synthesizes evidence from multiple biomedical sources, assesses interactions between drugs and patient conditions, and refines treatment recommendations through iterative reasoning. It selects tools based on task objectives and executes structured function calls to solve therapeutic tasks that require clinical reasoning and cross-source validation. The ToolUniverse consolidates 211 tools from trusted sources, including all US FDA-approved drugs since 1939 and validated clinical insights from Open Targets. TxAgent outperforms leading LLMs, tool-use models, and reasoning agents across five new benchmarks: DrugPC, BrandPC, GenericPC, TreatmentPC, and DescriptionPC, covering 3,168 drug reasoning tasks and 456 personalized treatment scenarios. It achieves 92.1% accuracy in open-ended drug reasoning tasks, surpassing GPT-4o and outperforming DeepSeek-R1 (671B) in structured multi-step reasoning. TxAgent generalizes across drug name variants and descriptions. By integrating multi-step inference, real-time knowledge grounding, and tool-assisted decision-making, TxAgent ensures that treatment recommendations align with established clinical guidelines and real-world evidence, reducing the risk of adverse events and improving therapeutic decision-making.
Multimodal Medical Code Tokenizer
Feb 06, 2025



Abstract:Foundation models trained on patient electronic health records (EHRs) require tokenizing medical data into sequences of discrete vocabulary items. Existing tokenizers treat medical codes from EHRs as isolated textual tokens. However, each medical code is defined by its textual description, its position in ontological hierarchies, and its relationships to other codes, such as disease co-occurrences and drug-treatment associations. Medical vocabularies contain more than 600,000 codes with critical information for clinical reasoning. We introduce MedTok, a multimodal medical code tokenizer that uses the text descriptions and relational context of codes. MedTok processes text using a language model encoder and encodes the relational structure with a graph encoder. It then quantizes both modalities into a unified token space, preserving modality-specific and cross-modality information. We integrate MedTok into five EHR models and evaluate it on operational and clinical tasks across in-patient and out-patient datasets, including outcome prediction, diagnosis classification, drug recommendation, and risk stratification. Swapping standard EHR tokenizers with MedTok improves AUPRC across all EHR models, by 4.10% on MIMIC-III, 4.78% on MIMIC-IV, and 11.30% on EHRShot, with the largest gains in drug recommendation. Beyond EHR modeling, we demonstrate using MedTok tokenizer with medical QA systems. Our results demonstrate the potential of MedTok as a unified tokenizer for medical codes, improving tokenization for medical foundation models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge