Ayush Noori
Department of Biomedical Informatics, Harvard Medical School, Boston, MA, USA, Department of Engineering Science, University of Oxford, Oxford, UK
Autonomous Knowledge Graph Exploration with Adaptive Breadth-Depth Retrieval
Jan 20, 2026Abstract:Retrieving evidence for language model queries from knowledge graphs requires balancing broad search across the graph with multi-hop traversal to follow relational links. Similarity-based retrievers provide coverage but remain shallow, whereas traversal-based methods rely on selecting seed nodes to start exploration, which can fail when queries span multiple entities and relations. We introduce ARK: Adaptive Retriever of Knowledge, an agentic KG retriever that gives a language model control over this breadth-depth tradeoff using a two-operation toolset: global lexical search over node descriptors and one-hop neighborhood exploration that composes into multi-hop traversal. ARK alternates between breadth-oriented discovery and depth-oriented expansion without depending on a fragile seed selection, a pre-set hop depth, or requiring retrieval training. ARK adapts tool use to queries, using global search for language-heavy queries and neighborhood exploration for relation-heavy queries. On STaRK, ARK reaches 59.1% average Hit@1 and 67.4 average MRR, improving average Hit@1 by up to 31.4% and average MRR by up to 28.0% over retrieval-based and agentic training-free methods. Finally, we distill ARK's tool-use trajectories from a large teacher into an 8B model via label-free imitation, improving Hit@1 by +7.0, +26.6, and +13.5 absolute points over the base 8B model on AMAZON, MAG, and PRIME datasets, respectively, while retaining up to 98.5% of the teacher's Hit@1 rate.
Graph AI generates neurological hypotheses validated in molecular, organoid, and clinical systems
Dec 13, 2025Abstract:Neurological diseases are the leading global cause of disability, yet most lack disease-modifying treatments. We present PROTON, a heterogeneous graph transformer that generates testable hypotheses across molecular, organoid, and clinical systems. To evaluate PROTON, we apply it to Parkinson's disease (PD), bipolar disorder (BD), and Alzheimer's disease (AD). In PD, PROTON linked genetic risk loci to genes essential for dopaminergic neuron survival and predicted pesticides toxic to patient-derived neurons, including the insecticide endosulfan, which ranked within the top 1.29% of predictions. In silico screens performed by PROTON reproduced six genome-wide $α$-synuclein experiments, including a split-ubiquitin yeast two-hybrid system (normalized enrichment score [NES] = 2.30, FDR-adjusted $p < 1 \times 10^{-4}$), an ascorbate peroxidase proximity labeling assay (NES = 2.16, FDR $< 1 \times 10^{-4}$), and a high-depth targeted exome sequencing study in 496 synucleinopathy patients (NES = 2.13, FDR $< 1 \times 10^{-4}$). In BD, PROTON predicted calcitriol as a candidate drug that reversed proteomic alterations observed in cortical organoids derived from BD patients. In AD, we evaluated PROTON predictions in health records from $n = 610,524$ patients at Mass General Brigham, confirming that five PROTON-predicted drugs were associated with reduced seven-year dementia risk (minimum hazard ratio = 0.63, 95% CI: 0.53-0.75, $p < 1 \times 10^{-7}$). PROTON generated neurological hypotheses that were evaluated across molecular, organoid, and clinical systems, defining a path for AI-driven discovery in neurological disease.
TxAgent: An AI Agent for Therapeutic Reasoning Across a Universe of Tools
Mar 14, 2025Abstract:Precision therapeutics require multimodal adaptive models that generate personalized treatment recommendations. We introduce TxAgent, an AI agent that leverages multi-step reasoning and real-time biomedical knowledge retrieval across a toolbox of 211 tools to analyze drug interactions, contraindications, and patient-specific treatment strategies. TxAgent evaluates how drugs interact at molecular, pharmacokinetic, and clinical levels, identifies contraindications based on patient comorbidities and concurrent medications, and tailors treatment strategies to individual patient characteristics. It retrieves and synthesizes evidence from multiple biomedical sources, assesses interactions between drugs and patient conditions, and refines treatment recommendations through iterative reasoning. It selects tools based on task objectives and executes structured function calls to solve therapeutic tasks that require clinical reasoning and cross-source validation. The ToolUniverse consolidates 211 tools from trusted sources, including all US FDA-approved drugs since 1939 and validated clinical insights from Open Targets. TxAgent outperforms leading LLMs, tool-use models, and reasoning agents across five new benchmarks: DrugPC, BrandPC, GenericPC, TreatmentPC, and DescriptionPC, covering 3,168 drug reasoning tasks and 456 personalized treatment scenarios. It achieves 92.1% accuracy in open-ended drug reasoning tasks, surpassing GPT-4o and outperforming DeepSeek-R1 (671B) in structured multi-step reasoning. TxAgent generalizes across drug name variants and descriptions. By integrating multi-step inference, real-time knowledge grounding, and tool-assisted decision-making, TxAgent ensures that treatment recommendations align with established clinical guidelines and real-world evidence, reducing the risk of adverse events and improving therapeutic decision-making.
Multi Scale Graph Neural Network for Alzheimer's Disease
Nov 16, 2024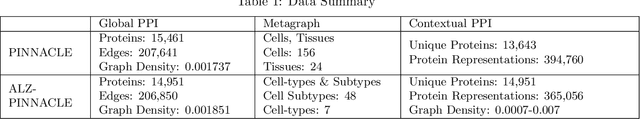
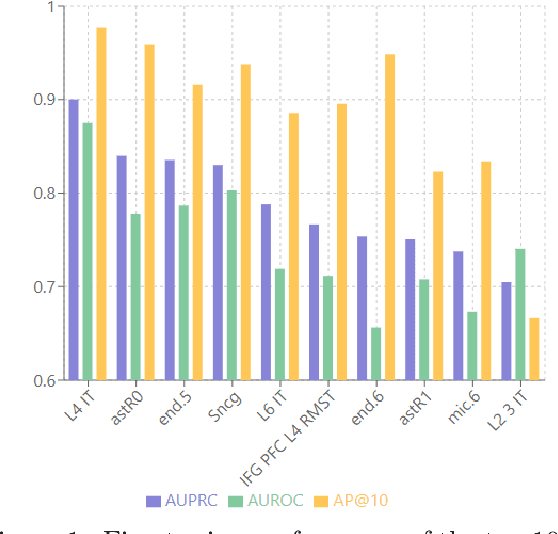
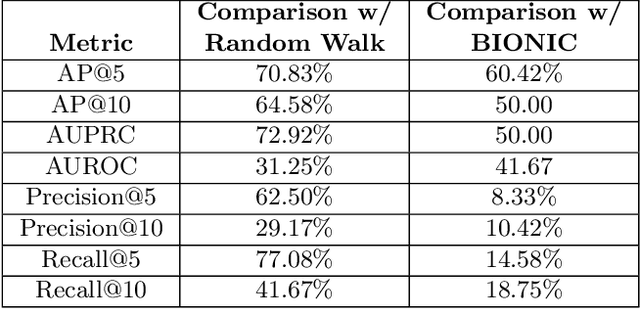
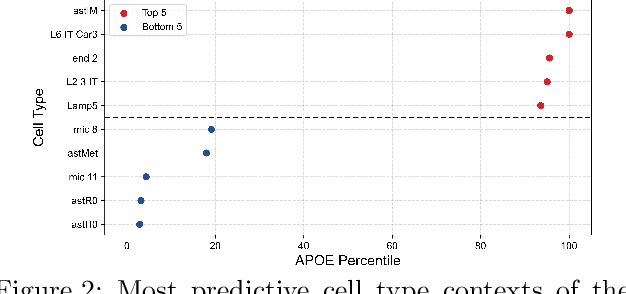
Abstract:Alzheimer's disease (AD) is a complex, progressive neurodegenerative disorder characterized by extracellular A\b{eta} plaques, neurofibrillary tau tangles, glial activation, and neuronal degeneration, involving multiple cell types and pathways. Current models often overlook the cellular context of these pathways. To address this, we developed a multiscale graph neural network (GNN) model, ALZ PINNACLE, using brain omics data from donors spanning the entire aging to AD spectrum. ALZ PINNACLE is based on the PINNACLE GNN framework, which learns context-aware protein, cell type, and tissue representations within a unified latent space. ALZ PINNACLE was trained on 14,951 proteins, 206,850 protein interactions, 7 cell types, and 48 cell subtypes or states. After pretraining, we investigated the learned embedding of APOE, the largest genetic risk factor for AD, across different cell types. Notably, APOE embeddings showed high similarity in microglial, neuronal, and CD8 cells, suggesting a similar role of APOE in these cell types. Fine tuning the model on AD risk genes revealed cell type contexts predictive of the role of APOE in AD. Our results suggest that ALZ PINNACLE may provide a valuable framework for uncovering novel insights into AD neurobiology.
Empowering Biomedical Discovery with AI Agents
Apr 03, 2024



Abstract:We envision 'AI scientists' as systems capable of skeptical learning and reasoning that empower biomedical research through collaborative agents that integrate machine learning tools with experimental platforms. Rather than taking humans out of the discovery process, biomedical AI agents combine human creativity and expertise with AI's ability to analyze large datasets, navigate hypothesis spaces, and execute repetitive tasks. AI agents are proficient in a variety of tasks, including self-assessment and planning of discovery workflows. These agents use large language models and generative models to feature structured memory for continual learning and use machine learning tools to incorporate scientific knowledge, biological principles, and theories. AI agents can impact areas ranging from hybrid cell simulation, programmable control of phenotypes, and the design of cellular circuits to the development of new therapies.
Graph AI in Medicine
Oct 20, 2023Abstract:In clinical artificial intelligence (AI), graph representation learning, mainly through graph neural networks (GNNs), stands out for its capability to capture intricate relationships within structured clinical datasets. With diverse data -- from patient records to imaging -- GNNs process data holistically by viewing modalities as nodes interconnected by their relationships. Graph AI facilitates model transfer across clinical tasks, enabling models to generalize across patient populations without additional parameters or minimal re-training. However, the importance of human-centered design and model interpretability in clinical decision-making cannot be overstated. Since graph AI models capture information through localized neural transformations defined on graph relationships, they offer both an opportunity and a challenge in elucidating model rationale. Knowledge graphs can enhance interpretability by aligning model-driven insights with medical knowledge. Emerging graph models integrate diverse data modalities through pre-training, facilitate interactive feedback loops, and foster human-AI collaboration, paving the way to clinically meaningful predictions.
Geometric multimodal representation learning
Sep 07, 2022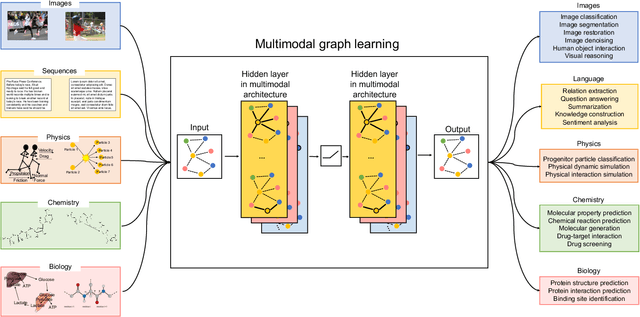
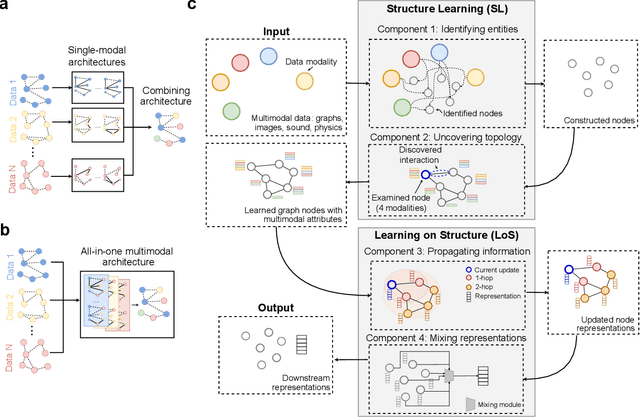
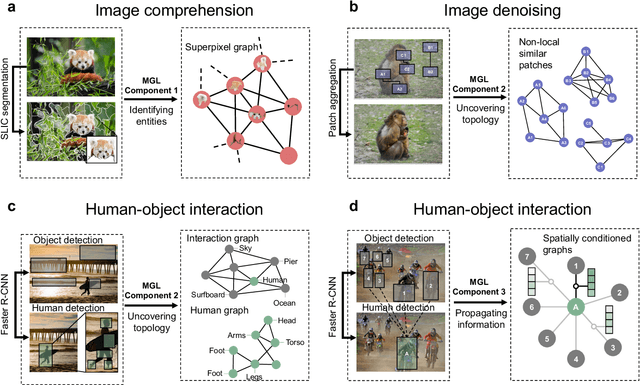

Abstract:Graph-centric artificial intelligence (graph AI) has achieved remarkable success in modeling interacting systems prevalent in nature, from dynamical systems in biology to particle physics. The increasing heterogeneity of data calls for graph neural architectures that can combine multiple inductive biases. However, combining data from various sources is challenging because appropriate inductive bias may vary by data modality. Multimodal learning methods fuse multiple data modalities while leveraging cross-modal dependencies to address this challenge. Here, we survey 140 studies in graph-centric AI and realize that diverse data types are increasingly brought together using graphs and fed into sophisticated multimodal models. These models stratify into image-, language-, and knowledge-grounded multimodal learning. We put forward an algorithmic blueprint for multimodal graph learning based on this categorization. The blueprint serves as a way to group state-of-the-art architectures that treat multimodal data by choosing appropriately four different components. This effort can pave the way for standardizing the design of sophisticated multimodal architectures for highly complex real-world problems.
Using Deep Learning to Identify Patients with Cognitive Impairment in Electronic Health Records
Nov 13, 2021


Abstract:Dementia is a neurodegenerative disorder that causes cognitive decline and affects more than 50 million people worldwide. Dementia is under-diagnosed by healthcare professionals - only one in four people who suffer from dementia are diagnosed. Even when a diagnosis is made, it may not be entered as a structured International Classification of Diseases (ICD) diagnosis code in a patient's charts. Information relevant to cognitive impairment (CI) is often found within electronic health records (EHR), but manual review of clinician notes by experts is both time consuming and often prone to errors. Automated mining of these notes presents an opportunity to label patients with cognitive impairment in EHR data. We developed natural language processing (NLP) tools to identify patients with cognitive impairment and demonstrate that linguistic context enhances performance for the cognitive impairment classification task. We fine-tuned our attention based deep learning model, which can learn from complex language structures, and substantially improved accuracy (0.93) relative to a baseline NLP model (0.84). Further, we show that deep learning NLP can successfully identify dementia patients without dementia-related ICD codes or medications.
Natural Language Processing to Detect Cognitive Concerns in Electronic Health Records Using Deep Learning
Nov 12, 2020
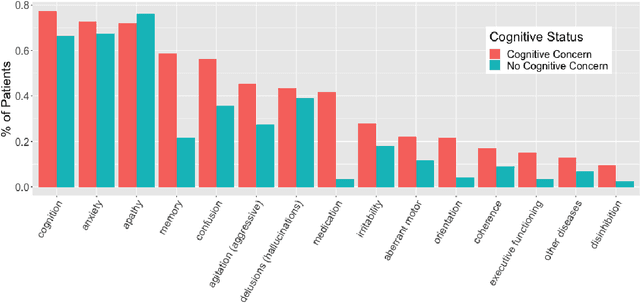
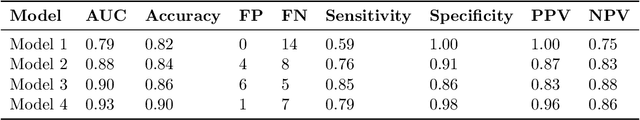
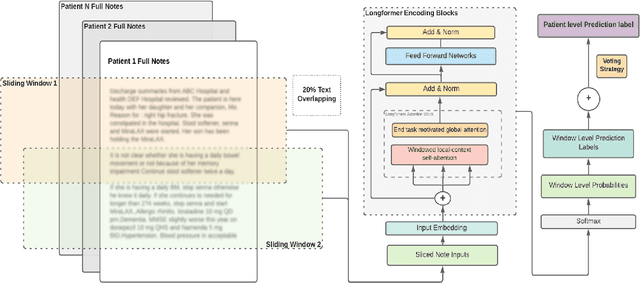
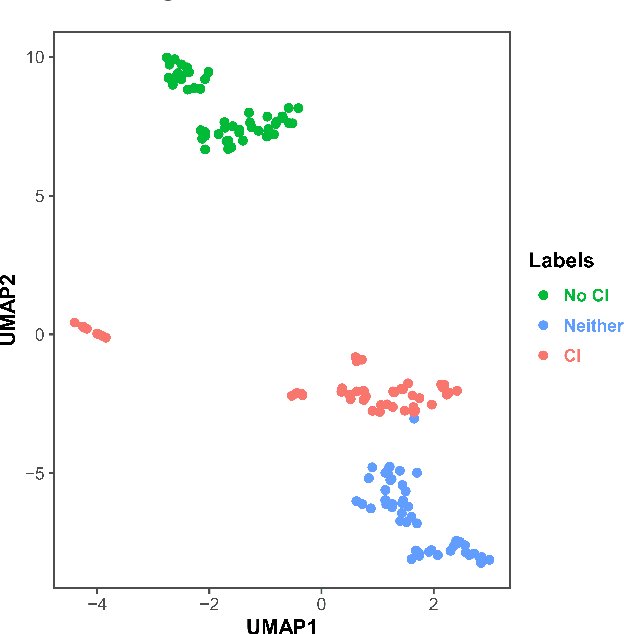
Abstract:Dementia is under-recognized in the community, under-diagnosed by healthcare professionals, and under-coded in claims data. Information on cognitive dysfunction, however, is often found in unstructured clinician notes within medical records but manual review by experts is time consuming and often prone to errors. Automated mining of these notes presents a potential opportunity to label patients with cognitive concerns who could benefit from an evaluation or be referred to specialist care. In order to identify patients with cognitive concerns in electronic medical records, we applied natural language processing (NLP) algorithms and compared model performance to a baseline model that used structured diagnosis codes and medication data only. An attention-based deep learning model outperformed the baseline model and other simpler models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge