Haoqi Sun
Bridging Large Language Models and Single-Cell Transcriptomics in Dissecting Selective Motor Neuron Vulnerability
May 12, 2025Abstract:Understanding cell identity and function through single-cell level sequencing data remains a key challenge in computational biology. We present a novel framework that leverages gene-specific textual annotations from the NCBI Gene database to generate biologically contextualized cell embeddings. For each cell in a single-cell RNA sequencing (scRNA-seq) dataset, we rank genes by expression level, retrieve their NCBI Gene descriptions, and transform these descriptions into vector embedding representations using large language models (LLMs). The models used include OpenAI text-embedding-ada-002, text-embedding-3-small, and text-embedding-3-large (Jan 2024), as well as domain-specific models BioBERT and SciBERT. Embeddings are computed via an expression-weighted average across the top N most highly expressed genes in each cell, providing a compact, semantically rich representation. This multimodal strategy bridges structured biological data with state-of-the-art language modeling, enabling more interpretable downstream applications such as cell-type clustering, cell vulnerability dissection, and trajectory inference.
Annotation of Sleep Depth Index with Scalable Deep Learning Yields Novel Digital Biomarkers for Sleep Health
Jul 05, 2024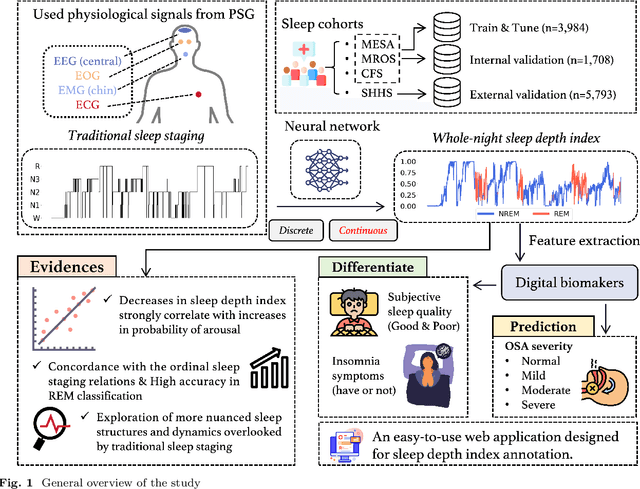

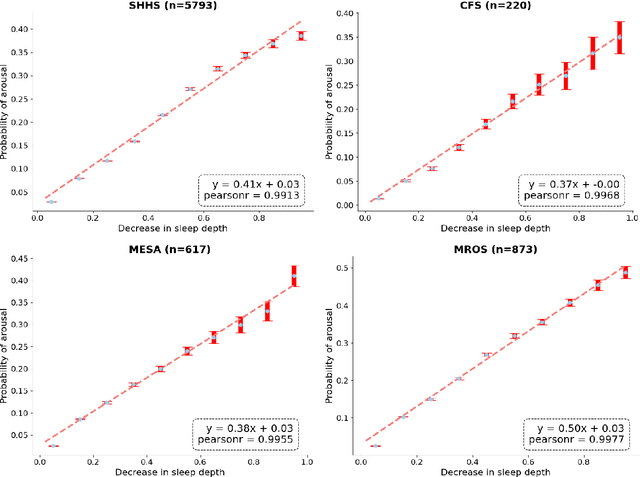
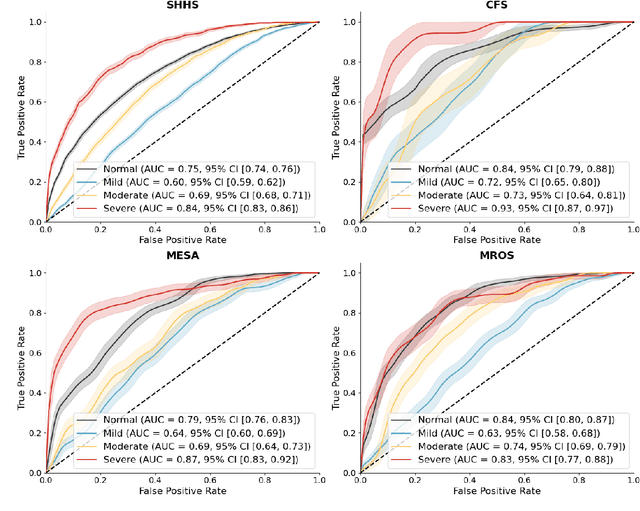
Abstract:Traditional sleep staging categorizes sleep and wakefulness into five coarse-grained classes, overlooking subtle variations within each stage. It provides limited information about the probability of arousal and may hinder the diagnosis of sleep disorders, such as insomnia. To address this issue, we propose a deep-learning method for automatic and scalable annotation of sleep depth index using existing sleep staging labels. Our approach is validated using polysomnography from over ten thousand recordings across four large-scale cohorts. The results show a strong correlation between the decrease in sleep depth index and the increase in arousal likelihood. Several case studies indicate that the sleep depth index captures more nuanced sleep structures than conventional sleep staging. Sleep biomarkers extracted from the whole-night sleep depth index exhibit statistically significant differences with medium-to-large effect sizes across groups of varied subjective sleep quality and insomnia symptoms. These sleep biomarkers also promise utility in predicting the severity of obstructive sleep apnea, particularly in severe cases. Our study underscores the utility of the proposed method for continuous sleep depth annotation, which could reveal more detailed structures and dynamics within whole-night sleep and yield novel digital biomarkers beneficial for sleep health.
Contactless Polysomnography: What Radio Waves Tell Us about Sleep
May 20, 2024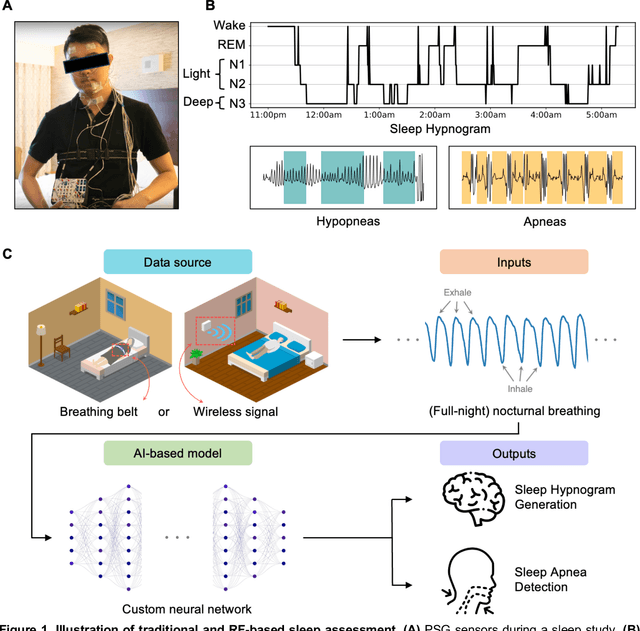
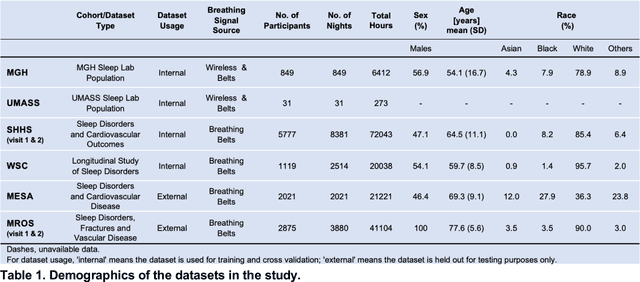
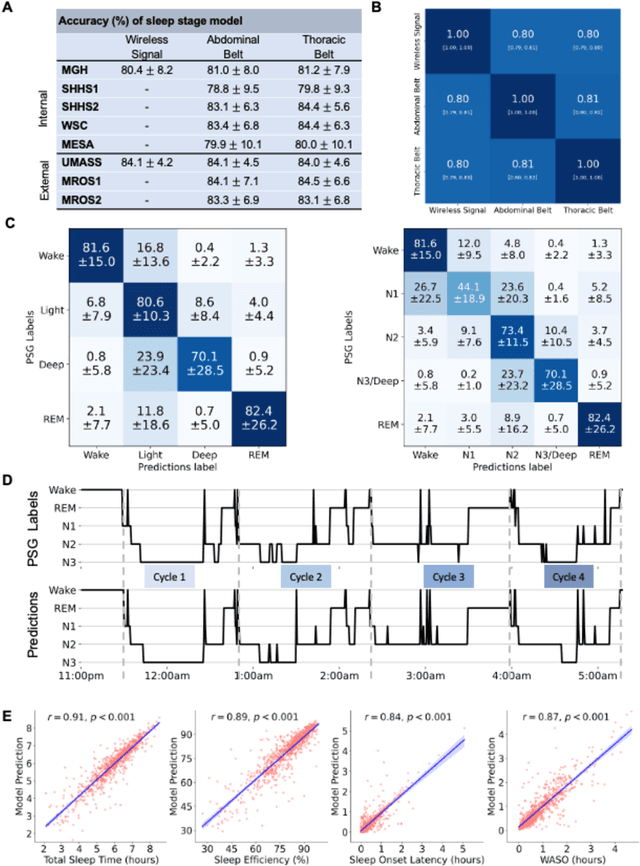
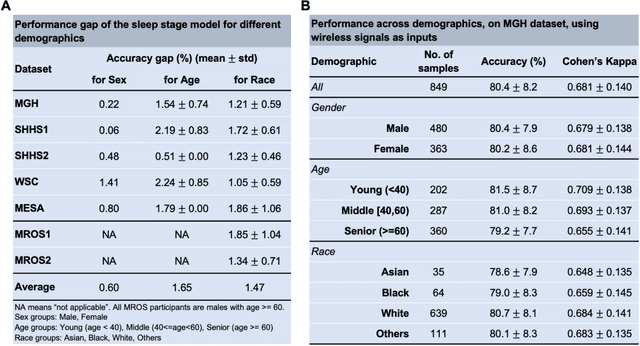
Abstract:The ability to assess sleep at home, capture sleep stages, and detect the occurrence of apnea (without on-body sensors) simply by analyzing the radio waves bouncing off people's bodies while they sleep is quite powerful. Such a capability would allow for longitudinal data collection in patients' homes, informing our understanding of sleep and its interaction with various diseases and their therapeutic responses, both in clinical trials and routine care. In this article, we develop an advanced machine learning algorithm for passively monitoring sleep and nocturnal breathing from radio waves reflected off people while asleep. Validation results in comparison with the gold standard (i.e., polysomnography) (n=849) demonstrate that the model captures the sleep hypnogram (with an accuracy of 81% for 30-second epochs categorized into Wake, Light Sleep, Deep Sleep, or REM), detects sleep apnea (AUROC = 0.88), and measures the patient's Apnea-Hypopnea Index (ICC=0.95; 95% CI = [0.93, 0.97]). Notably, the model exhibits equitable performance across race, sex, and age. Moreover, the model uncovers informative interactions between sleep stages and a range of diseases including neurological, psychiatric, cardiovascular, and immunological disorders. These findings not only hold promise for clinical practice and interventional trials but also underscore the significance of sleep as a fundamental component in understanding and managing various diseases.
Why Interpretable Causal Inference is Important for High-Stakes Decision Making for Critically Ill Patients and How To Do It
Mar 09, 2022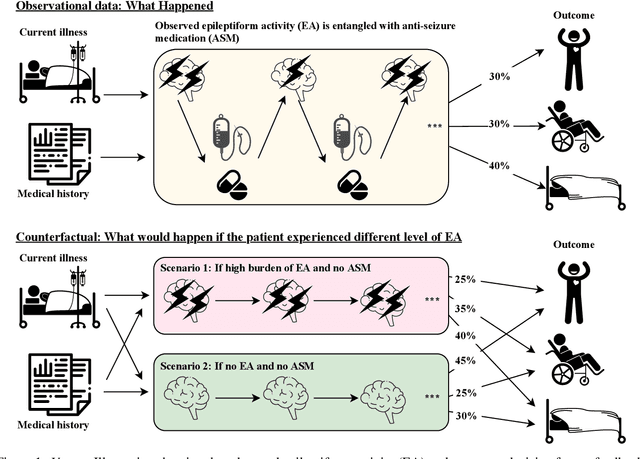
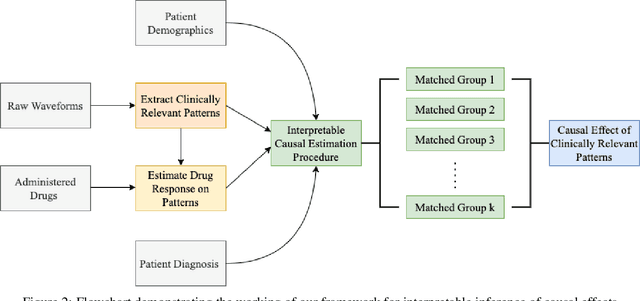
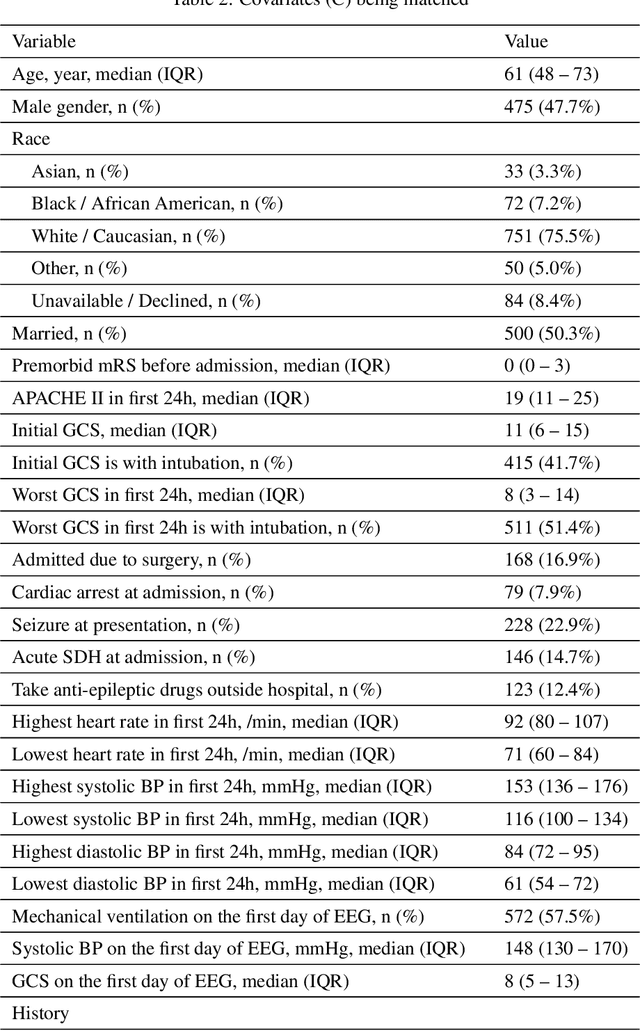
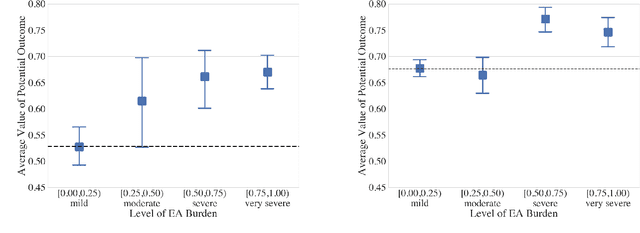
Abstract:Many fundamental problems affecting the care of critically ill patients lead to similar analytical challenges: physicians cannot easily estimate the effects of at-risk medical conditions or treatments because the causal effects of medical conditions and drugs are entangled. They also cannot easily perform studies: there are not enough high-quality data for high-dimensional observational causal inference, and RCTs often cannot ethically be conducted. However, mechanistic knowledge is available, including how drugs are absorbed into the body, and the combination of this knowledge with the limited data could potentially suffice -- if we knew how to combine them. In this work, we present a framework for interpretable estimation of causal effects for critically ill patients under exactly these complex conditions: interactions between drugs and observations over time, patient data sets that are not large, and mechanistic knowledge that can substitute for lack of data. We apply this framework to an extremely important problem affecting critically ill patients, namely the effect of seizures and other potentially harmful electrical events in the brain (called epileptiform activity -- EA) on outcomes. Given the high stakes involved and the high noise in the data, interpretability is critical for troubleshooting such complex problems. Interpretability of our matched groups allowed neurologists to perform chart reviews to verify the quality of our causal analysis. For instance, our work indicates that a patient who experiences a high level of seizure-like activity (75% high EA burden) and is untreated for a six-hour window, has, on average, a 16.7% increased chance of adverse outcomes such as severe brain damage, lifetime disability, or death. We find that patients with mild but long-lasting EA (average EA burden >= 50%) have their risk of an adverse outcome increased by 11.2%.
Sleep Apnea and Respiratory Anomaly Detection from a Wearable Band and Oxygen Saturation
Feb 24, 2021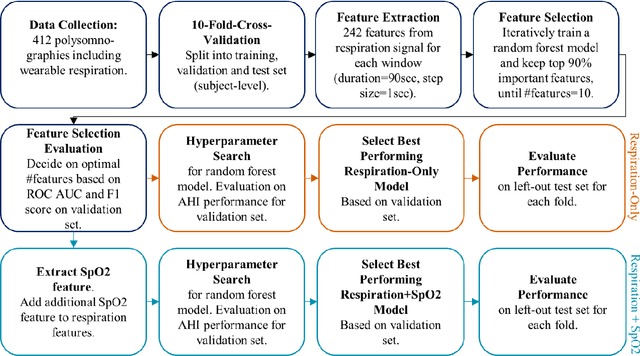
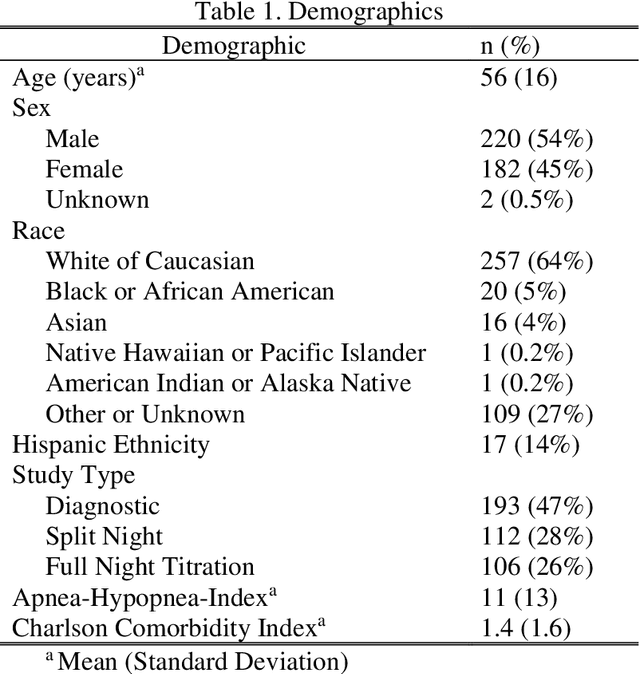
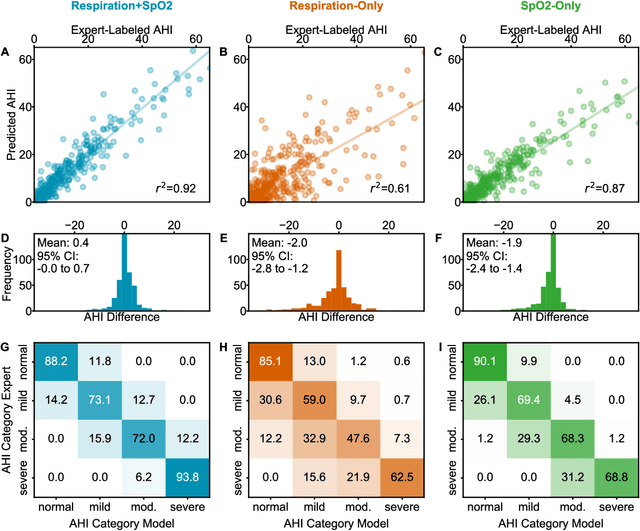
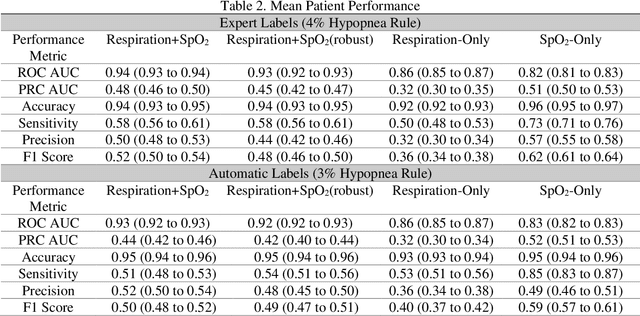
Abstract:Objective: Sleep related respiratory abnormalities are typically detected using polysomnography. There is a need in general medicine and critical care for a more convenient method to automatically detect sleep apnea from a simple, easy-to-wear device. The objective is to automatically detect abnormal respiration and estimate the Apnea-Hypopnea-Index (AHI) with a wearable respiratory device, compared to an SpO2 signal or polysomnography using a large (n = 412) dataset serving as ground truth. Methods: Simultaneously recorded polysomnographic (PSG) and wearable respiratory effort data were used to train and evaluate models in a cross-validation fashion. Time domain and complexity features were extracted, important features were identified, and a random forest model employed to detect events and predict AHI. Four models were trained: one each using the respiratory features only, a feature from the SpO2 (%)-signal only, and two additional models that use the respiratory features and the SpO2 (%)-feature, one allowing a time lag of 30 seconds between the two signals. Results: Event-based classification resulted in areas under the receiver operating characteristic curves of 0.94, 0.86, 0.82, and areas under the precision-recall curves of 0.48, 0.32, 0.51 for the models using respiration and SpO2, respiration-only, and SpO2-only respectively. Correlation between expert-labelled and predicted AHI was 0.96, 0.78, and 0.93, respectively. Conclusions: A wearable respiratory effort signal with or without SpO2 predicted AHI accurately. Given the large dataset and rigorous testing design, we expect our models are generalizable to evaluating respiration in a variety of environments, such as at home and in critical care.
Automated Respiratory Event Detection Using Deep Neural Networks
Jan 12, 2021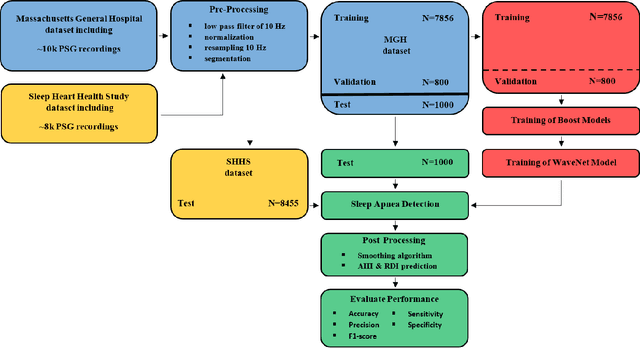
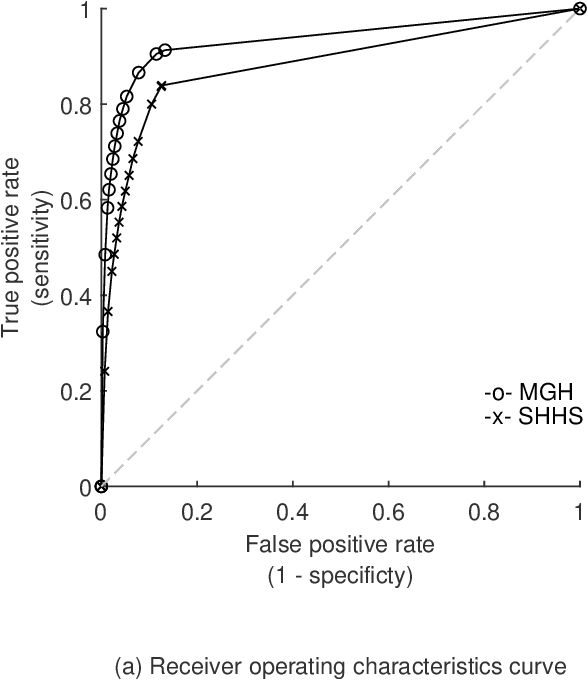
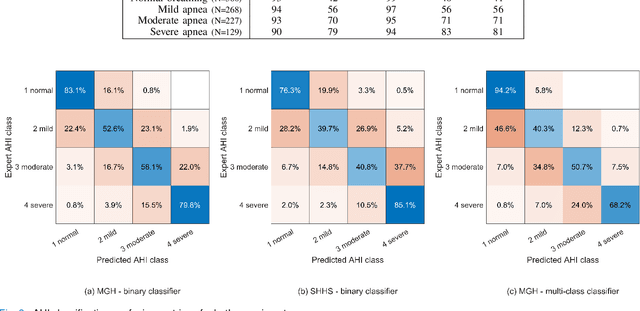
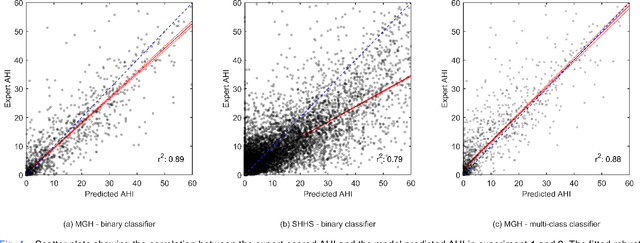
Abstract:The gold standard to assess respiration during sleep is polysomnography; a technique that is burdensome, expensive (both in analysis time and measurement costs), and difficult to repeat. Automation of respiratory analysis can improve test efficiency and enable accessible implementation opportunities worldwide. Using 9,656 polysomnography recordings from the Massachusetts General Hospital (MGH), we trained a neural network (WaveNet) based on a single respiratory effort belt to detect obstructive apnea, central apnea, hypopnea and respiratory-effort related arousals. Performance evaluation included event-based and recording-based metrics - using an apnea-hypopnea index analysis. The model was further evaluated on a public dataset, the Sleep-Heart-Health-Study-1, containing 8,455 polysomnographic recordings. For binary apnea event detection in the MGH dataset, the neural network obtained an accuracy of 95%, an apnea-hypopnea index $r^2$ of 0.89 and area under the curve for the receiver operating characteristics curve and precision-recall curve of 0.93 and 0.74, respectively. For the multiclass task, we obtained varying performances: 81% of all labeled central apneas were correctly classified, whereas this metric was 46% for obstructive apneas, 29% for respiratory effort related arousals and 16% for hypopneas. The majority of false predictions were misclassifications as another type of respiratory event. Our fully automated method can detect respiratory events and assess the apnea-hypopnea index with sufficient accuracy for clinical utilization. Differentiation of event types is more difficult and may reflect in part the complexity of human respiratory output and some degree of arbitrariness in the clinical thresholds and criteria used during manual annotation.
Natural Language Processing to Detect Cognitive Concerns in Electronic Health Records Using Deep Learning
Nov 12, 2020
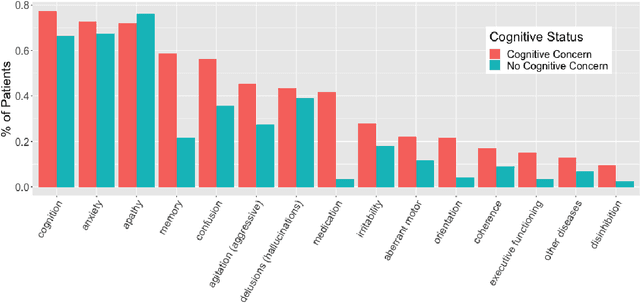
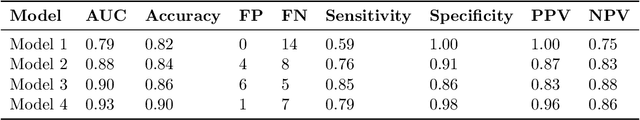
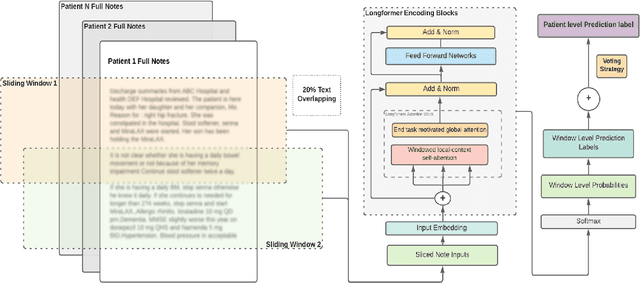
Abstract:Dementia is under-recognized in the community, under-diagnosed by healthcare professionals, and under-coded in claims data. Information on cognitive dysfunction, however, is often found in unstructured clinician notes within medical records but manual review by experts is time consuming and often prone to errors. Automated mining of these notes presents a potential opportunity to label patients with cognitive concerns who could benefit from an evaluation or be referred to specialist care. In order to identify patients with cognitive concerns in electronic medical records, we applied natural language processing (NLP) algorithms and compared model performance to a baseline model that used structured diagnosis codes and medication data only. An attention-based deep learning model outperformed the baseline model and other simpler models.
HAMLET: Interpretable Human And Machine co-LEarning Technique
Aug 21, 2018



Abstract:Efficient label acquisition processes are key to obtaining robust classifiers. However, data labeling is often challenging and subject to high levels of label noise. This can arise even when classification targets are well defined, if instances to be labeled are more difficult than the prototypes used to define the class, leading to disagreements among the expert community. Here, we enable efficient training of deep neural networks. From low-confidence labels, we iteratively improve their quality by simultaneous learning of machines and experts. We call it Human And Machine co-LEarning Technique (HAMLET). Throughout the process, experts become more consistent, while the algorithm provides them with explainable feedback for confirmation. HAMLET uses a neural embedding function and a memory module filled with diverse reference embeddings from different classes. Its output includes classification labels and highly relevant reference embeddings as explanation. We took the study of brain monitoring at intensive care unit (ICU) as an application of HAMLET on continuous electroencephalography (cEEG) data. Although cEEG monitoring yields large volumes of data, labeling costs and difficulty make it hard to build a classifier. Additionally, while experts agree on the labels of clear-cut examples of cEEG patterns, labeling many real-world cEEG data can be extremely challenging. Thus, a large minority of sequences might be mislabeled. HAMLET has shown significant performance gain against deep learning and other baselines, increasing accuracy from 7.03% to 68.75% on challenging inputs. Besides improved performance, clinical experts confirmed the interpretability of those reference embeddings in helping explaining the classification results by HAMLET.
SLEEPNET: Automated Sleep Staging System via Deep Learning
Jul 26, 2017



Abstract:Sleep disorders, such as sleep apnea, parasomnias, and hypersomnia, affect 50-70 million adults in the United States (Hillman et al., 2006). Overnight polysomnography (PSG), including brain monitoring using electroencephalography (EEG), is a central component of the diagnostic evaluation for sleep disorders. While PSG is conventionally performed by trained technologists, the recent rise of powerful neural network learning algorithms combined with large physiological datasets offers the possibility of automation, potentially making expert-level sleep analysis more widely available. We propose SLEEPNET (Sleep EEG neural network), a deployed annotation tool for sleep staging. SLEEPNET uses a deep recurrent neural network trained on the largest sleep physiology database assembled to date, consisting of PSGs from over 10,000 patients from the Massachusetts General Hospital (MGH) Sleep Laboratory. SLEEPNET achieves human-level annotation performance on an independent test set of 1,000 EEGs, with an average accuracy of 85.76% and algorithm-expert inter-rater agreement (IRA) of kappa = 79.46%, comparable to expert-expert IRA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge